Interleukin 8 protein promotes inflammatory responses, even in airways. The presence of interleukin 8 gene variants causes altered inflammatory responses and possibly varied responses to inhaled bronchodilators. Thus, this study analyzed the interleukin 8 variants (rs4073, rs2227306, and rs2227307) and their association with the response to inhaled bronchodilators in cystic fibrosis patients.
MethodsAnalysis of interleukin 8 gene variants was performed by restriction fragment length polymorphism of polymerase chain reaction. The association between spirometry markers and the response to inhaled bronchodilators was evaluated by Mann–Whitney and Kruskal–Wallis tests. The analysis included all cystic fibrosis patients, and subsequently patients with two mutations in the cystic fibrosis transmembrane conductance regulator gene belonging to classes I to III.
ResultsThis study included 186 cystic fibrosis patients. There was no association of the rs2227307 variant with the response to inhaled bronchodilators. The rs2227306 variant was associated with FEF50% in the dominant group and in the group with two identified mutations in the cystic fibrosis transmembrane conductance regulator gene. The rs4073 variant was associated with spirometry markers in four genetic models: co-dominant (FEF25–75% and FEF75%), dominant (FEV1, FEF50%, FEF75%, and FEF25–75%), recessive (FEF75% and FEF25–75%), and over-dominant (FEV1/FVC).
ConclusionsThis study highlighted the importance of the rs4073 variant of the interleukin 8 gene, regarding response to inhaled bronchodilators, and of the assessment of mutations in the cystic fibrosis transmembrane conductance regulator gene.
A proteína interleucina 8 promove respostas inflamatórias, o que inclui sua atuação nas vias aéreas. A presença de variantes no gene da interleucina 8 causa respostas inflamatórias alteradas e possivelmente respostas variadas ao uso de broncodilatadores inalatórios. Assim, este estudo analisou as variantes da interleucina 8 (rs4073, rs2227306, rs2227307) e sua associação à resposta a broncodilatadores inalatórios em pacientes com fibrose cística.
MétodosFoi feita análise das variantes genéticas da interleucina 8 por restriction fragment length polymorphism da reação em cadeia da polimerase. A associação entre os marcadores da espirometria e a resposta a broncodilatadores inalatórios foi feita pelos testes de Mann-Whitney e Kruskal-Wallis. A análise incluiu todos os pacientes com fibrose cística e posteriormente pacientes com duas mutações no gene cystic fibrosis transmembrane conductance regulator pertencentes às Classes I a II.
ResultadosEste estudo incluiu 186 pacientes com fibrose cística. Não houve associação da variante rs2227307 à resposta a broncodilatadores inalatórios. A variante rs2227306 foi associada a FEF50% no grupo dominante e no grupo com duas mutações identificadas no gene cystic fibrosis transmembrane conductance regulator. A variante rs4073 foi associada a marcadores da espirometria em quatro modelos genéticos: codominante (FEF25-75% e FEF75%), dominante (VEF1, FEF50%, FEF75% e FEF25-75%), recessivo (FEF75% e FEF25-75%) e overdominante (VEF1/CVF).
ConclusõesEste estudo destaca, principalmente, a importância da variante rs4073 do gene da interleucina 8, na resposta a broncodilatadores inalatórios, concomitantemente ao genótipo das mutações no gene cystic fibrosis transmembrane conductance regulator.
The response to inhaled bronchodilators (BD) in cystic fibrosis (CF) (OMIM: n. 219700) is quite variable and depends on the cystic fibrosis transmembrane regulator (CFTR) genotype, pulmonary symptoms, and mainly, on variants in the modifier genes, such as the beta-2-adrenergic receptor (ADRB2).1 So far, only a few studies have investigated such response.
Mutations in the CFTR gene cause CF, due to deficiency, dysfunction, or absence of the CFTR protein.2 CF is characterized by a continuous cycle of chronic airway inflammation, which can be exacerbated by interleukin-8 (IL-8), a key pro-inflammatory mediator. IL-8 is responsible for initiating and increasing the inflammatory response in the presence of specific pathogens, causing activation and migration of neutrophils from peripheral blood to tissues.3 Chronic airway inflammation is the final common pathway of lung injury. It is responsible for increased vascular permeability, contributing to interstitial, alveolar, and airway edema.
The treatment of CF lung disease includes anti-inflammatory drugs, inhaled corticosteroids, antibiotics, mucolytic, hypertonic saline, and physiotherapy. Among the possible therapies, there is little evidence supporting the role of BD in CF.4,5 However, BD is often prescribed for a longer period due to wheezing and dyspnea episodes in CF.6 BD reduces the liberation of mediators, which are responsible for recruiting and activating inflammatory cells, activating cholinergic neurotransmission and improving vascular permeability. It also increases mucociliary clearance, leading to reduced lung inflammation.7 The response to BD depends, in part, on the CFTR protein, which interacts with the ADRB2 protein to promote broncodilatation.8 Spirometry is the primary method to assess lung function, severity and progression of the disease, as well as response to BD.
Intense neutrophil inflammation and low bronchial hyperresponsiveness are commonly observed characteristics in CF.9 Moreover, the response of genetic variants to BD is little known.1 The role of IL-8 in the neutrophilic component of CF lung disease can be pointed out. Genes that might be associated with the severity of CF and possibly with the response to BD have been reported in the present study and in the literature.1,10–12 The IL-8 gene should be highlighted, as it has a direct influence on lung inflammation and can be effective in the response to BD. Therefore, this study compared the rs4073, rs2227306, and rs2227307 IL-8 gene variants with the response to BD in CF patients, using spirometry.
MethodsPatientsA cross-sectional study was conducted in 186 CF patients, selected in two university referral centers (Medical School of São José do Rio Preto and Universidade Estadual de Campinas) for CF treatment, from 2013 to 2015. The study was approved by the Research Ethics Committee of the Universidade Estadual de Campinas, Brazil (under No. 528/2008). All participants were informed of the study and signed an informed consent form. For patients under the age of 18, the informed consent form was signed by the parents or guardians. The study followed the recommendations of the Declaration of Helsinki.
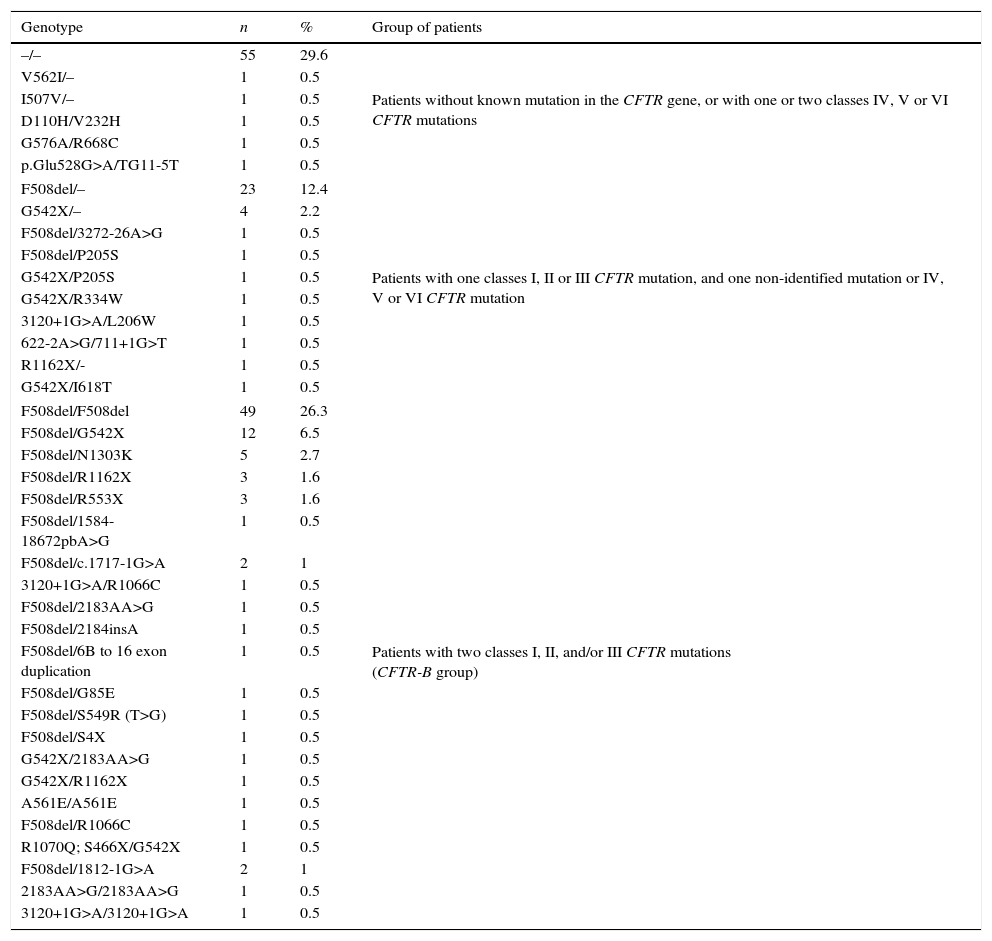
The diagnosis of CF was confirmed by the presence of two altered concentrations of sodium and chloride in sweat (chloride level higher than 60mEq/L). Furthermore, no patient underwent initial immunoreactive trypsinogen (IRT) measurement. In a group of 91 patients, there were two mutations in the CFTR gene belonging to classes I, II, and/or III (associated with greater disease severity, due to the absence or non-functionality of the CFTR protein)13; 60 patients did not present an identified mutation of the CFTR gene, or had two mutations belonging to classes IV, V, or VI; and 35 patients had a mutation in the CFTR gene belonging to classes I, II, or III, and a non-identified mutation, or belonging to classes IV, V, or VI (Table 1).
Distribution of cystic fibrosis patients for the CFTR genotype and classes of identified mutations.a
| Genotype | n | % | Group of patients |
|---|---|---|---|
| –/– | 55 | 29.6 | Patients without known mutation in the CFTR gene, or with one or two classes IV, V or VI CFTR mutations |
| V562I/– | 1 | 0.5 | |
| I507V/– | 1 | 0.5 | |
| D110H/V232H | 1 | 0.5 | |
| G576A/R668C | 1 | 0.5 | |
| p.Glu528G>A/TG11-5T | 1 | 0.5 | |
| F508del/– | 23 | 12.4 | Patients with one classes I, II or III CFTR mutation, and one non-identified mutation or IV, V or VI CFTR mutation |
| G542X/– | 4 | 2.2 | |
| F508del/3272-26A>G | 1 | 0.5 | |
| F508del/P205S | 1 | 0.5 | |
| G542X/P205S | 1 | 0.5 | |
| G542X/R334W | 1 | 0.5 | |
| 3120+1G>A/L206W | 1 | 0.5 | |
| 622-2A>G/711+1G>T | 1 | 0.5 | |
| R1162X/- | 1 | 0.5 | |
| G542X/I618T | 1 | 0.5 | |
| F508del/F508del | 49 | 26.3 | Patients with two classes I, II, and/or III CFTR mutations (CFTR-B group) |
| F508del/G542X | 12 | 6.5 | |
| F508del/N1303K | 5 | 2.7 | |
| F508del/R1162X | 3 | 1.6 | |
| F508del/R553X | 3 | 1.6 | |
| F508del/1584-18672pbA>G | 1 | 0.5 | |
| F508del/c.1717-1G>A | 2 | 1 | |
| 3120+1G>A/R1066C | 1 | 0.5 | |
| F508del/2183AA>G | 1 | 0.5 | |
| F508del/2184insA | 1 | 0.5 | |
| F508del/6B to 16 exon duplication | 1 | 0.5 | |
| F508del/G85E | 1 | 0.5 | |
| F508del/S549R (T>G) | 1 | 0.5 | |
| F508del/S4X | 1 | 0.5 | |
| G542X/2183AA>G | 1 | 0.5 | |
| G542X/R1162X | 1 | 0.5 | |
| A561E/A561E | 1 | 0.5 | |
| F508del/R1066C | 1 | 0.5 | |
| R1070Q; S466X/G542X | 1 | 0.5 | |
| F508del/1812-1G>A | 2 | 1 | |
| 2183AA>G/2183AA>G | 1 | 0.5 | |
| 3120+1G>A/3120+1G>A | 1 | 0.5 | |
n, sample size; CFTR, cystic fibrosis transmembrane regulator.
The following variables were analyzed: clinical scores (Shwachman-Kulczycki, Kanga, and Bhalla); body mass index (BMI) for patients older than 18 years, using the formula BMI=weight/(height)2, the Anthro program version 3.0.1 (World Health Organization [WHO], 2006) was used for children under five years of age and the Anthro Plus version 1.0.2 (World Health Organization [WHO], 2007) was used for patients aged between five and 18 years; patient's age and age at diagnosis; first clinical symptom (one general symptom, pulmonary and digestive symptoms); period until the first colonization with Pseudomonas aeruginosa; microorganisms identified in the routine sputum culture (mucoid and non-mucoid P. aeruginosa, Achromobacter xylosoxidans, Burkolderia cepacia, and Staphylococcus aureus); transcutaneous arterial hemoglobin oxygen-saturation (SaO2); spirometry; and comorbidities (nasal polyps, osteoporosis, meconium ileus, diabetes mellitus, and pancreatic insufficiency).
Spirometry was performed in patients over seven years of age, using the CPFS/D spirometer (MedGraphics – Saint Paul, Minnesota, USA). Data were recorded in the PF BREEZE software version 3.8B for Windows 95/98/NT (American Thoracic Society), and assessed in percentage predicted values for: forced vital capacity (FVC), forced expiratory volume in 1s (FEV1), FEV1/FVC ratio, forced expiratory flow at 25% of FVC (FEF25%), forced expiratory flow at 50% of FVC (FEF50%), forced expiratory flow at 75% of FVC (FEF75%), forced expiratory flow between 25% and 75% of FVC (FEF25–75%), maximum forced expiratory flow (FEFmax), and expiratory reserve volume (ERV). Spirometry data were shown in percentage of the predicted value according to the Polgar and Promadhat (1971), Pereira et al. (2007), and Duarte et al. (2007) equations.14–16
In all patients undergoing spirometry, the test was performed before and 15min after administration of BD (albuterol – C13H21NO3 [400mg]). Patients under BD therapy were instructed to interrupt the medication eight hours prior to spirometry, if they were under short-acting BD treatment; and 48hours, if they were under long-acting BD treatment. The post-BD percentage change was used for the statistical analysis. The BD response criteria, defined as an increase of >12% and 200mL of initial FEV1, was used as a second model to evaluate the association between the IL-8 gene variants and BD response.
DNA extraction and genotypingGenomic DNA was extracted from peripheral blood samples using standard phenol-chloroform method and quantified by GE NanoVue™ spectrophotometer (GE Healthcare Biosciences – Pittsburgh, USA). In this study, the final sample concentration was set at 50ng/μL.
Mutations of the CFTR gene were analyzed by polymerase chain reaction (PCR; F508del) followed by enzymatic digestion (G542X, R1162X, R553X, G551D, and N1303K). Other mutations in the CFTR gene were identified by sequencing or with the use of the SALSA Multiplex Ligation-dependent Probe Amplification (MPLA method) Kit P091-C1 CFTR-MRC-Holland S4X, 2183A>G, 1717-G>A, I618T with MegaBace1000® (GE Healthcare Biosciences – Pittsburgh, USA), and ABI3500 (Applied Biosystems – Thermo Fisher Scientific – São Paulo, Brazil).17
IL-8 gene variants were analyzed by PCR followed by restriction enzyme digestion. For the rs4073 variant, the primers 5′-CCA TCA TGA TAG CAT CTG TA-3′ and 5′-CCA CAA TTT GGT GAA TTA TTA A-3′, and the AseI restriction enzyme were used; for the rs2227306 variant, primers 5′-CTC TAA CTC TTT ATA TAG GAA TT-3′ and 5′-GAT TGA TTT TAT CAA CAG GCA-3′, as well as the EcoRI restriction enzyme; and for the rs2227307 variant, primers 5′-TAA AGG TTT GAT CAA TAT AGA-3′ and 5′-CTT CCT TCT AAT TCCA ATA TG-3′, as well as the ScrFI restriction enzyme.18,19 The products of the enzymatic restriction were submitted to electrophoresis on a 12% polyacrylamide gel, or 4% agarose gel,18,19 and stained with Red® gel.
Statistical analysisStatistical analyses were performed using Statistical Package for the Social Sciences version 22.0 (SPSS Inc. – Chicago, USA). The GPower software version 3.1.9.220 was used to calculate the sample power, considering the genotype of the analyzed variants and adopting power for value above 80%. The following conditions were applied to calculate the sample power: analysis of variance test, in place of Kruskal–Wallis test considering that analysis of variance is a stronger test (effect size=0.25, α=0.05, power=0.80, numerator degree of freedom=2, number of groups=3, ideal n=158); two-tailed Mann–Whitney test (effect size=0.5, α=0.05, power=0.80, allocation ratio N2/N1=1, ideal n=134); chi-squared test (effect size=0.3, α=0.05, power=0.80, degree of freedom=2, ideal n=108).
The Mann–Whitney and Kruskal–Wallis tests were used for the comparison between different genotypes and groups of IL-8 gene variants and the response to BD. In case of significant differences between the groups for the Kruskal–Wallis test, further identification and evaluation of the differences between genotypes were performed with MedCalc® software for Windows, version 16.1 (MedCalc® Software – Ostend, Belgium).
The chi-squared test and Fisher's exact test were used for the comparison between different genotypes and groups of IL-8 gene variants, as well as the response to BD, defined as an increase of >12% and 200mL of initial FEV1.
For the identification of mutations in the CFTR gene, patients were analyzed based on two contexts: CFTR-A, all CF patients, regardless of gene mutations (n=186 patients); and CFTR-B, patients with two mutations belonging to classes I, II, and/or III (n=91 patients). As for the variants, four analysis models were adopted: (i) co-dominant (Kruskal–Wallis test); (ii) recessive (Mann–Whitney test); (iii) dominant (Mann–Whitney test); (iv) over-dominant (Mann–Whitney test), applied in association with clinical variables. For all analyses, the alpha value was set at 0.05.
For analysis of the Hardy–Weinberg equilibrium (HWE), the Online Encyclopedia software for Genetic Epidemiology Studies (OEGE) was used.
The false discovery rate (FDR) test was applied to correct the multiple test comparison. FDR is an approach to the multiple comparisons problem. Instead of controlling the chance of any false, FDR controls the expected proportion of false positives among supra threshold voxels. A FDR threshold is determined from the observed p-value distribution, and hence is adaptive to the amount of signal in the data.21 The p-value and corrected p-value (pc) were shown in the manuscript. The linkage disequilibrium analysis was performed in Haploview software version 4.2.
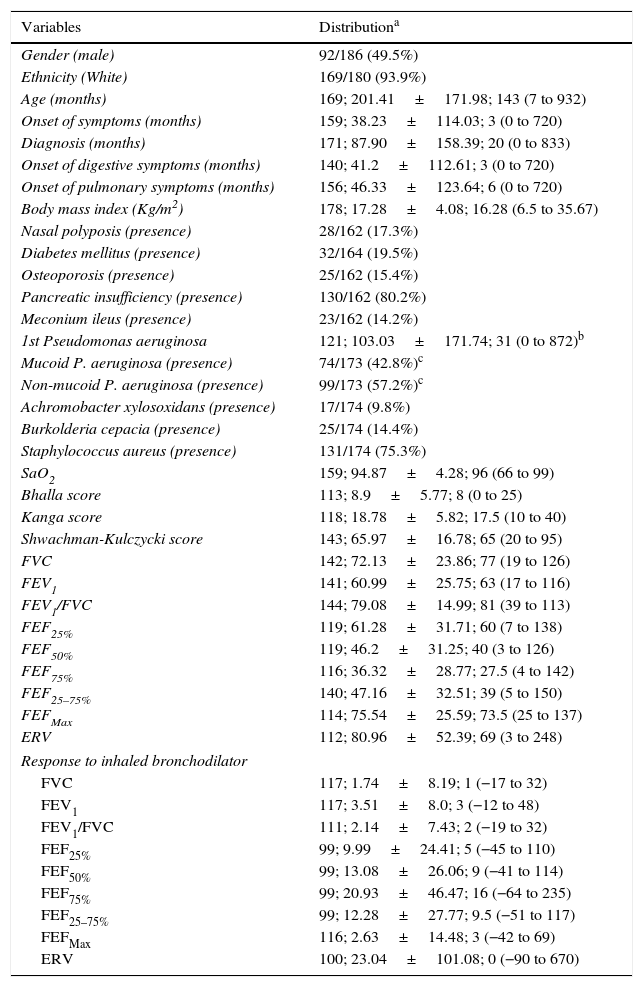
ResultsClinical and laboratory data of CF patients are described in Table 2. Table 3 describes the genotype and allele frequency of IL-8 gene variants. Fig. 1 shows the data with p-values and corrected p-values for association of the three IL-8 gene variants, considering the four models of analysis proposed and the genotype of the CFTR gene.
Descriptive analysis of clinical and laboratory markers of cystic fibrosis patients.
| Variables | Distributiona |
|---|---|
| Gender (male) | 92/186 (49.5%) |
| Ethnicity (White) | 169/180 (93.9%) |
| Age (months) | 169; 201.41±171.98; 143 (7 to 932) |
| Onset of symptoms (months) | 159; 38.23±114.03; 3 (0 to 720) |
| Diagnosis (months) | 171; 87.90±158.39; 20 (0 to 833) |
| Onset of digestive symptoms (months) | 140; 41.2±112.61; 3 (0 to 720) |
| Onset of pulmonary symptoms (months) | 156; 46.33±123.64; 6 (0 to 720) |
| Body mass index (Kg/m2) | 178; 17.28±4.08; 16.28 (6.5 to 35.67) |
| Nasal polyposis (presence) | 28/162 (17.3%) |
| Diabetes mellitus (presence) | 32/164 (19.5%) |
| Osteoporosis (presence) | 25/162 (15.4%) |
| Pancreatic insufficiency (presence) | 130/162 (80.2%) |
| Meconium ileus (presence) | 23/162 (14.2%) |
| 1st Pseudomonas aeruginosa | 121; 103.03±171.74; 31 (0 to 872)b |
| Mucoid P. aeruginosa (presence) | 74/173 (42.8%)c |
| Non-mucoid P. aeruginosa (presence) | 99/173 (57.2%)c |
| Achromobacter xylosoxidans (presence) | 17/174 (9.8%) |
| Burkolderia cepacia (presence) | 25/174 (14.4%) |
| Staphylococcus aureus (presence) | 131/174 (75.3%) |
| SaO2 | 159; 94.87±4.28; 96 (66 to 99) |
| Bhalla score | 113; 8.9±5.77; 8 (0 to 25) |
| Kanga score | 118; 18.78±5.82; 17.5 (10 to 40) |
| Shwachman-Kulczycki score | 143; 65.97±16.78; 65 (20 to 95) |
| FVC | 142; 72.13±23.86; 77 (19 to 126) |
| FEV1 | 141; 60.99±25.75; 63 (17 to 116) |
| FEV1/FVC | 144; 79.08±14.99; 81 (39 to 113) |
| FEF25% | 119; 61.28±31.71; 60 (7 to 138) |
| FEF50% | 119; 46.2±31.25; 40 (3 to 126) |
| FEF75% | 116; 36.32±28.77; 27.5 (4 to 142) |
| FEF25–75% | 140; 47.16±32.51; 39 (5 to 150) |
| FEFMax | 114; 75.54±25.59; 73.5 (25 to 137) |
| ERV | 112; 80.96±52.39; 69 (3 to 248) |
| Response to inhaled bronchodilator | |
| FVC | 117; 1.74±8.19; 1 (−17 to 32) |
| FEV1 | 117; 3.51±8.0; 3 (−12 to 48) |
| FEV1/FVC | 111; 2.14±7.43; 2 (−19 to 32) |
| FEF25% | 99; 9.99±24.41; 5 (−45 to 110) |
| FEF50% | 99; 13.08±26.06; 9 (−41 to 114) |
| FEF75% | 99; 20.93±46.47; 16 (−64 to 235) |
| FEF25–75% | 99; 12.28±27.77; 9.5 (−51 to 117) |
| FEFMax | 116; 2.63±14.48; 3 (−42 to 69) |
| ERV | 100; 23.04±101.08; 0 (−90 to 670) |
SaO2, transcutaneous arterial hemoglobin oxygen-saturation; FVC, forced vital capacity; FEV1, forced expiratory volume in 1s of FVC; FEF25%, forced expiratory flow at 25% of FVC; FEF50%, forced expiratory flow at 50% of FVC; FEF75%, forced expiratory flow at 75% of FVC; FEF25–75%, average forced expiratory flow between 25% and 75% of FVC; FEFmax, maximum forced expiratory flow; ERV, expiratory reserve volume. Spirometry data are shown in percentage of the predicted value. All evaluated patients are described.
The data with categorical distribution are presented as follows: n of variable/n total (percentage); data with numeric distribution are presented as follows: sample size; mean±standard deviation; median (minimum to maximum).
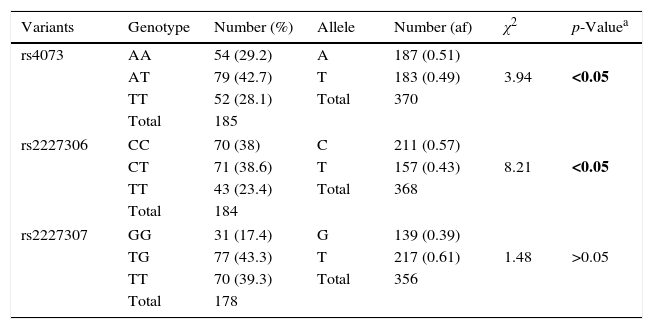
Distribution of genotypes, alleles and haplotype of IL-8 gene variants (rs4073, rs2227306 and rs2227307) in cystic fibrosis patients.
| Variants | Genotype | Number (%) | Allele | Number (af) | χ2 | p-Valuea |
|---|---|---|---|---|---|---|
| rs4073 | AA | 54 (29.2) | A | 187 (0.51) | 3.94 | <0.05 |
| AT | 79 (42.7) | T | 183 (0.49) | |||
| TT | 52 (28.1) | Total | 370 | |||
| Total | 185 | |||||
| rs2227306 | CC | 70 (38) | C | 211 (0.57) | 8.21 | <0.05 |
| CT | 71 (38.6) | T | 157 (0.43) | |||
| TT | 43 (23.4) | Total | 368 | |||
| Total | 184 | |||||
| rs2227307 | GG | 31 (17.4) | G | 139 (0.39) | 1.48 | >0.05 |
| TG | 77 (43.3) | T | 217 (0.61) | |||
| TT | 70 (39.3) | Total | 356 | |||
| Total | 178 | |||||
| rs4073/rs2227306/rs2227307 | Frequency | Percentage (%) |
|---|---|---|
| AA CC GG | 3 | 1.7 |
| AA CC GT | 5 | 2.8 |
| AA CC TT | 24 | 13.6 |
| AA CT GG | 3 | 1.7 |
| AA CT GT | 4 | 2.3 |
| AA CT TT | 7 | 4 |
| AA TT GG | 2 | 1.1 |
| AA TT GT | 2 | 1.1 |
| AA TT TT | 3 | 1.7 |
| AT CC GT | 13 | 7.3 |
| AT CC TT | 7 | 4 |
| AT CT GT | 30 | 16.9 |
| AT CT TT | 10 | 5.6 |
| AT TT GG | 5 | 2.8 |
| AT TT GT | 6 | 3.4 |
| AT TT TT | 3 | 1.7 |
| TT CC GG | 2 | 1.1 |
| TT CC GT | 5 | 2.8 |
| TT CC TT | 8 | 4.5 |
| TT CT GG | 4 | 2.3 |
| TT CT GT | 6 | 3.4 |
| TT CT TT | 3 | 1.7 |
| TT TT GG | 12 | 6.8 |
| TT TT GT | 5 | 2.8 |
| TT TT TT | 5 | 2.8 |
| Total | 177 | 100 |
IL-8, interleukin 8; %, percentage; χ2, chi-square; af, absolute frequency.
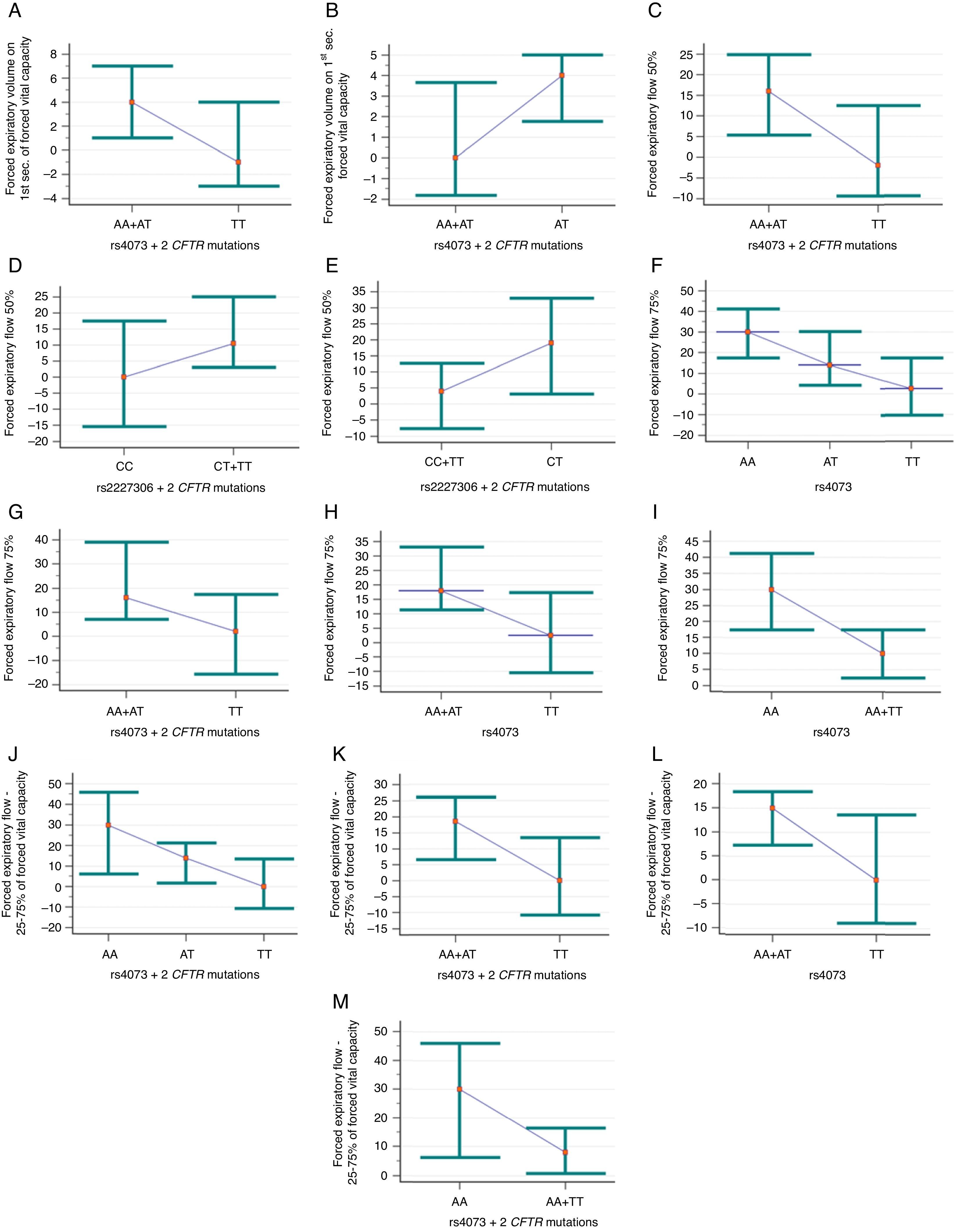
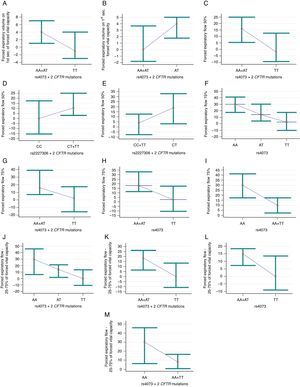
Association of rs4073 and rs2227306 variants of IL-8 (interleukin-8) with the response to inhaled bronchodilators in cystic fibrosis patients. (A) Association between forced expiratory volume in 1s (FEV1) of forced vital capacity (FVC) and rs4073, dominant model and two identified mutations in the CFTR gene (cystic fibrosis transmembrane regulator) belonging to classes I, II, and/or III (CFTR-B group) (p=0.028; pc=0.112). (AA+AT) n=39; mean of 4.77±8.95; median of 4 (ranging from −9 to 48). (TT) n=21; mean of −0.86±5.42; median of −1 (ranging from −6 to 15). (B) Association of FEV1/FVC ratio with rs4073, over-dominant model and CFTR-B group (p=0.029; pc=0.116). (AA+TT) n=30; mean of 1.17±7.36; median of 0 (ranging from −12 to 23). (AT) n=29; mean of 4.76±7.58; median of 4 (ranging from −9 to 32). (C) Association of forced expiratory flow of 50% (FEF50%) of FVC with rs4073, dominant model, and CFTR-B group (p=0.046; pc=0.184). (AA+AT) n=37; mean of 17.38±24.18; median of 16 (ranging from −20 to 89). (TT) n=21; mean of 4.95±20.8; median of −2 (ranging from −19 to 55). (D) Association of FEF50% with rs2227306, dominant model, and CFTR-B group (p=0.05; pc=0.083). (CC) n=15; mean of 1.47±17.22; median of 0 (ranging from −20 to 29). (CT+TT) n=44; mean of 15.84±25.04; median of 10.5 (ranging from −28 to 89). (E) Association of FEF50% with rs2227306, over-dominant model, and CFTR-B group (p=0.033; pc=0.083). (CC+TT) n=33; mean of 6.06±20.27; median of 4 (ranging from −20 to 55). (CT) n=23; mean of 19.96±26.43; median of 19 (ranging from −28 to 89). (F) Association of forced expiratory flow of 75% (FEF75%) of FVC with rs4073, co-dominant model regardless of identified mutations in the CFTR gene (CFTR-A group) (p=0.044; pc=0.058). 1≠3. (AA) n=28; mean of 29.93±39.98; median of 30 (ranging from −47 to 142). (AT) n=42; mean of 24.81±53.57; median of 14 (ranging from −58 to 235). (TT) n=28; mean of 9.14±36.95; median of 2.5 (ranging from −35 to 119). (G) Association of FEF75% with rs4073, dominant model, and CFTR-B group (p=0.024; pc=0.096). (AA+AT) n=38; mean of 33±55.23; median of 16 (ranging from −27 to 235). (TT) n=21; mean of 5.43±35.93; median of 2 (ranging from −35 to 119). (H) Association of FEF75% with rs4073, dominant model, and CFTR-A group (p=0.034; pc=0.058). (AA+AT) n=70; mean of 26.86±48.34; median of 18 (ranging from −58 to 235). (TT) n=28; mean of 9.14±36.95; median of 2.5 (ranging from −35 to 119). (I) Association of FEF75% with rs4073, recessive model, and CFTR-A group (p=0.04; pc=0.058). (AA) n=28; mean of 29.93±39.98; median of 30 (ranging from −47 to 142). (AT+TT) n=70; mean of 18.54±47.95; median of 10 (ranging from −58 to 235). (J) Association of FEF between 25% and 75% (FEF25–75%) of FVC with rs4073, co-dominant model and CFTR-B group (p=0.012; pc=0.024). TT≠AA and AT. (AA) n=9; mean of 25.78±23.14; median of 30 (ranging from −21 to 57). (AT) n=29; mean of 18.38±29.04; median of 14 (ranging from −33 to 117). (TT) n=21; mean of 2.14±23.74; median of 0 (ranging from −51 to 65). (K) Association of FEF25–75% with rs4073, dominant model, and CFTR-B group (p=0.007; pc=0.024). (AA+ATT) n=38; mean of 20.13±27.64; median of 18.5 (ranging from −33 to 117). (TT) n=23; mean of 2.14±23.74; median of 0 (ranging from −51 to 65). (L) Association of FEF25–75% with rs4073, dominant model, and CFTR-A group (p=0.029; pc=0.1). (AA+AT) n=82; mean of 15.48±27.2; median of 15 (ranging from −40 to 117). (TT) n=33; mean of 5.94±27.33; median of 0 (ranging from −51 to 76). (M) Association of FEF25–75% with rs4073, recessive model, and CFTR-B group (p=0.047; pc=0.063). (AA) n=9; mean of 25.78±23.14; median of 30 (ranging from −21 to 57). (AT+TT) n=50; mean of 11.56±27.89; median of 8 (ranging from −51 to 117). The dot corresponds to median values and the bar corresponds to the 95% confidence interval.
The rs2227307 variant was not associated with response to BD in any of the studied models; in turn, rs2227306 was associated with FEF50% for patients with CC genotype (dominant model; p=0.05; pc=0.083) and CT genotype (over-dominant model; p=0.033; pc=0.083) in the CFTR-B group (Fig. 1D and E).
Special emphasis should be given to the rs4073 variant, which was associated with the following spirometry variables: FEV1, FEV1/FVC ratio, FEF50% FEF75%, and FEF25–75%. The lowest response to BD was observed for the TT genotype (dominant model) and patients of the CFTR-B group to FEV1 (Fig. 1A), FEF50% (Fig. 1C), FEF75% (Fig. 1G), and FEF25–75% (Fig. 1K). The same was observed for the CFTR-A group for FEF75% (Fig. 1H) and FEF25–75% (Fig. 1L). In the co-dominant analysis, the TT genotype presented a lower response to BD for FEF75% (Fig. 1F) and FEF25–75% (Fig. 1J), respectively, in the CFTR-A and CFTR-B groups. For the marker FEV1/FVC, the AT genotype in patients from the CFTR-B group showed lower response to BD (Fig. 1B). Finally, in patients from the CFTR-A and CFTR-B groups the AA genotype (recessive model) for rs4073 showed higher response to BD for FEF75% (Fig. 1I) and FEF25–75%, respectively (Fig. 1M).
The haplotype distribution for the IL-8 gene variants is presented in Table 3.
For the comparison between IL-8 gene variants and the response to BD, defined as an increase of >12% and 200mL of initial FEV1, no positive association was found (p>0.05).
DiscussionThe influence of different IL-8 gene variants in the clinical severity of CF has been previously demonstrated.22 This study showed the association of different IL-8 gene variants and their modulation to BD response, assessing the impact of the drug on pulmonary function. The variability of responses to BD is determined by multiple factors, such as inflammation and pulmonary obstruction,23 bacteria,24 and lung symptoms,25 as well as modifier genes.26 However, no studies so far have investigated the role of IL-8 as a pro-inflammatory mediator of CF and its relationship to BD, and whether the IL-8 gene variants may explain the individual response to BD in CF. It is believed that the short-acting and long-acting beta-2-agonists may be beneficial for CF patients with positive bronchial hyperresponsiveness.6
In progressive lung diseases, different markers have been assessed; the use of BD appears to provide better response to FEF25–75%, when compared with other markers, which indicates involvement of smaller caliber airways in CF.27 In the present study, in agreement with the reference literature, an association was observed between the rs4073 variant and the FEF25–75% marker in the dominant (for both CFTR-A and CFTR-B groups), co-dominant (CFTR-B group), and recessive models (CFTR-B group). There was improvement in several other markers of spirometry for rs4073, such as FEV1, FEV1/FVC, FEF50%, and FEF75%, which shows impact on breathing patterns of patients.
The study by Hillian et al. (2008) found an association of rs2227307 and rs4073 IL-8 gene variants and severity of pulmonary disease.28 In that study, patients were divided into two cohorts: (1) homozygous F508del patients; (2) patients with other genotypes of the CFTR gene. In cohort 1, the rs4073, rs2227306, and rs2227543 variants were not associated with lung disease, and an association was observed for rs22227307, regardless of gender. In cohort 2, the rs4073 and rs2227306 variants were associated with lung disease severity in males. Thus, the gender of the patient, genotype of the CFTR gene, and modifier genes may modulate the severity of the lung disease.28 In this study, the rs2227307 genotype did not have an impact on the variability of the response to BD. This suggests that although this genotype is associated with lung disease in homozygous F508del patients, its response to BD is not relevant.
The target of BD is the ADRB2 protein, which is expressed in the airway smooth muscle. Variants of ADRB2 are associated with response to the medication. Although this protein has been widely studied in asthma, it was very seldom studied in CF. The effectiveness of the response to BD and inhaled corticosteroids to manage airway inflammation in asthma has been confirmed, and depends on the Arg16Gly (rs1042713; c.46A>G) and Glu27Gln (rs1042714; c.79C>G) ADRB2 gene variants.29 The Gln27Glu variant confers resistance to the ADRB2 protein in BD response. The 46*G and 79*G alleles are protective against asthma, reducing the risk by 27%.29 Both variants of the G allele induce changes in the regulation of the receptor due to increased susceptibility to protein degradation. This study found that the Arg16Gly and Gln27Glu ADRB2 gene variants influence the response to BD in CF. In spirometry and in other markers of severity, the Arg16Gly variant showed a positive association, unlike Gln27Glu. The Arg/Arg genotype for the Arg16Gly variant was associated with better values of FEV1 and FEF25–75%. The response to BD in the analyses of haplotype was positive in the absence of Gly16Gly and Glu27Glu genotypes for FEV1/FVC ratio.
Variants of ADRB2, diffusing capacity of the lungs for carbon monoxide, alveolar capillary membrane conductance, volume of blood in the alveolar capillary, and SaO2 were evaluated in the response to BD in 18 patients and 20 healthy controls, before and after the administration of salbutamol (30, 60, and 90min). Healthy subjects showed no changes in markers assessed for the Glu27Glu variant. However, in CF, this variant influenced the response to BD: the best response was found in the presence of at least one allele 27Glu. There was a difference in pulmonary diffusion and peripheral SaO2 according to the variation of the ADRB2 gene at position 27, and the dosage of the drug should be prescribed according to this variation.30
Most studies focus on the ADRB2 gene variants in the response to BD. However, this study demonstrated that, even indirectly, the IL-8 gene variants (and possibly in other genes, which modulate the inflammatory lung response) may potentiate or minimize the effect of BD and also influence the response to the medication.
Regarding the HWE, as previously discussed by this group,22 two variants (rs4073 and rs2227306) were not in balance. It is important to remember that the HWE assumes an ideal population, without the interference of evolutionary factors. However, in genes as those involved in immunity, inflammation, and infection control, the HWE imbalance may appear secondarily associated with the selection mechanisms that favored a particular allele that can bring a more effective response. The disequilibrium does not invalidate the association study since the groups are part of the same population.
The limitations of the present study include (i) cross-sectional dataset (BD response was evaluated at a single time); (ii) numerous missing data considering the problems to achieve information in the records; (iii) whether BD response differs based on baseline FEV1, mainly in health subjects; (iv) potential confounders to the results, such as disease severity, current therapies, medication adherence, and adequate pulmonary function test effort.
In conclusion, IL-8 gene variants (rs2227306 and specially rs4073) can be associated with the response to BD during spirometry. This drug may be an alternative for the treatment of the disease, mostly among patients with airways of smaller caliber. Studies involving dosage and combinations of drugs should be further conducted, in order determine the best treatment according to CF patient genotype, the CFTR gene, as well as modifier genes.
FundingFALM: Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) for supporting the researches #2011/12939-4, #2011/18845-1, #2015/12183-8, and #2015/12858-5; Fundo de Apoio à Pesquisa, ao Ensino e à Extensão da Universidade Estadual de Campinas for supporting the research #0648/2015; JDR: FAPESP for supporting the research #2011/18845-1 and #2015/12183-8. LLF: FAPESP for supporting the research #2013/19052-0.
Conflicts of interestThe authors declare no conflicts of interest.
To Luciana Montes Rezende, Luciana Cardoso Bonadia, and Stephanie Villa-Nova for their technical support during DNA extraction and identification of mutations of the CFTR gene. To Marcela Augusta de Souza Pinhel, Michele Lima Gregório, Rafael Fernandes Ferreira, Graciele Domitila Tenani, and Heloisa Cristina Caldas for their technical support during standardization of IL-8 genotyping. To Maria Ângela Gonçalves de Oliveira Ribeiro for conducting pulmonary function tests (LAFIP/Ciped/Unicamp). To Rafaella Maionchi Pereira Martins for her technical support to determine clinical scores. To Maria de Fátima Corrêa Pimenta Servidoni for promoting a link between both Universities.
Please cite this article as: Furlan LL, Ribeiro JD, Bertuzzo CS, Salomão Junior JB, Souza DR, Marson FA. Variants in the interleukin 8 gene and the response to inhaled bronchodilators in cystic fibrosis. J Pediatr (Rio J). 2017;93:639–48.
Study conducted at Universidade Estadual de Campinas (Unicamp), Campinas, SP, Brazil.