Obesity is a chronic disease caused by both environmental and genetic factors. Epidemiological studies have documented that increased energy intake and sedentary lifestyle, as well as a genetic contribution, are forces behind the obesity epidemic. Knowledge about the interaction between genetic and environmental components can facilitate the choice of the most effective and specific measures for the prevention of obesity. The aim of this study was to assess the association between the FTO, AKT1, and AKTIP genes and childhood obesity and insulin resistance.
MethodsThis was a case–control study in which SNPs in the FTO (rs99396096), AKT1, and AKTIP genes were genotyped in groups of controls and obese/overweight children. The study included 195 obese/overweight children and 153 control subjects.
ResultsAs expected, the obese/overweight group subjects had higher body mass index, higher fasting glucose, HOMA-IR index, total cholesterol, low-density lipoprotein, and triglycerides. However, no significant differences were observed in genes polymorphisms genotype or allele frequencies.
ConclusionThe present results suggest that AKT1, FTO, and AKTIP polymorphisms were not associated with obesity/overweight in Brazilians children. Future studies on the genetics of obesity in Brazilian children and their environment interactions are needed.
A obesidade é uma doença crônica sustentada por fatores ambientais e genéticos. Estudos epidemiológicos documentaram que maior ingestão de energia e um estilo de vida sedentário, bem como a contribuição genética, são forças por trás da epidemia de obesidade. O conhecimento sobre a interação entre os componentes genéticos e ambientais pode facilitar a escolha das medidas mais efetivas e específicas para a prevenção da obesidade. O objetivo deste estudo foi avaliar a relação entre os genes FTO, AKT1 e AKTIP e a obesidade infantil e resistência à insulina.
MétodosEstudo de caso-controle no qual SNPs nos genes FTO (rs99396096), AKT1 e AKTIP foram genotipados em grupos de controle e de crianças obesas/acima do peso. Foram recrutadas 195 crianças obesas/acima do peso e 153 indivíduos controle.
ResultadosComo esperado, os indivíduos do grupo obeso/acima do peso apresentaram maior índice de massa corporal, maior glicemia de jejum, índice de HOMA-IR, colesterol total, lipoproteína de baixa densidade e triglicerídeos. Contudo, não encontramos diferenças significativas no genótipo de polimorfismos gênicos ou nas frequências alélicas.
ConclusãoNossos resultados sugerem que os polimorfismos AKT1, FTO e AKTIP não estavam associados à obesidade/sobrepeso em crianças brasileiras. São necessários estudos futuros sobre a genética da obesidade em crianças brasileiras e suas interações ambientais.
Childhood obesity is a public health problem worldwide. Over the past decades, rates of overweight and obesity among children have largely increased both in developed and developing countries.1 Obesity is a result of environmental factors interacting with a polygenic background; the heritability ranges from 40% to 70%.2 A recent meta-regression analysis showed that heritability was higher in children than in adults.3 Long-term studies have shown that childhood obesity leads to clustering of metabolic syndrome (MetS) components,4 which include abdominal obesity, dyslipidemia, insulin resistance, type II diabetes, and hypertension.5 It is well known that overweight and obese children have a higher risk to become obese in adulthood.6
Genome-wide association studies (GWAS) identified the Fat mass and obesity associated gene (FTO) as associated with human adiposity.7 The FTO gene is related to obesity risk, especially the single nucleotide polymorphism (SNP) rs9939609, which has been further confirmed by others independent studies in different human populations.8–11 Due to the close relationship between diabetes and obesity, another interesting gene is the V-Akt murine thymoma viral oncogene homolog 1 (AKT1), which is thought to mediate many metabolic, mitogenic, and anti-apoptotic effects of insulin, IGF-1, and IL-3, and other growth factors and cytokines.12–14 Moreover, Akt also stimulates glucose uptake and glycogen synthesis,14 as well as protein synthesis.15 Several studies correlate insulin resistance to impairments at Akt pathway and, in certain conditions, these alterations can be of genetic origin. AKT1-binding protein (AKTIP or Ft1), located near to FTO at the same GWAS risk locus for obesity at chromosome 16 (16q12.2),16 appears to be another target for investigation. AKTIP, as a direct-ligand of AKT, modulates integration signaling pathways.17
Energy balance is influenced by a number of variables such as diet, social structures, metabolic factors, modern sedentary lifestyle, inexpensive energy-dense foods, and genetics. Probably, common obesity is the result of an adverse obesogenic environment and a susceptible genotype.4 To date, evidence for the possible clinical benefits of genetic studies for common complex diseases has been limited. Furthermore, the search for genetic factors involved in obesity is a challenge, and can provide extra data to answer such a complex question. Through these observations, this study aimed to investigate the statistical association of polymorphisms in the FTO, AKT1, and AKTIP genes with childhood obesity in Brazilian children.
MethodsClinical dataParents and children were informed and signed a written consent about the nature and purpose of this study. All children were submitted to a thorough physical examination. Height was measured using a wall-mounted stadiometer while body weight was measured through a digital scale. Body mass index (BMI) was calculated using weight and height measurements according to the equation BMI=weight (kg)/height2 (cm). The BMI cut-off points adopted were those established by the World Health Organization (WHO).18 Overweight and obesity were defined as BMI greater than +1 and +2 standard deviation, respectively.19 Blood pressure was measured at least twice and the presence of hypertension was defined as systolic and/or diastolic blood pressure above the 95th percentile.20 None of the children used medication.
Lipid profile and glycemia were determined in serum and plasma, respectively, with routine enzymatic commercial kits (Labtest Diagnóstica, S.A., Brazil). Insulin levels were measured using a kit (Genesis Diagnostics Products, São Paulo, Brazil). Insulin resistance was calculated by homeostasis model assessment as an index of insulin resistance (HOMA-IR), as described by Wallace et al.21 To date, few studies have defined cut-off levels for HOMA-IR in prepubertal and pubertal individuals. Based on that, HOMA-IR was tested as continuous variable and was controlled by the BMI status of the individuals.
The local ethics committee approved this study under the protocol CAAE No. 06400000180-7.
Overweight and obese patientsThe present sample included 195 obese/overweight subjects with mean age of 11.0 (±3.9; 104 females), with BMI 26.7 (±4.5). All subjects were recruited from the Endocrinology Ambulatory of the Adolescent and Child Institute and from the Childhood Endocrinology Ambulatory of the IMEPEN Foundation in Juiz de Fora, Brazil, and lived in urban areas.22,23
Control groupIndividuals in the control group were carefully selected from the local community and consisted of 153 eutrophic, normotensive individuals with mean age of 11.9 (±3.2; 91 females), and BMI 17.9 (±2.3). All subjects signed an informed consent form and the Research Ethics Committee of the University approved the protocols.
GenotypingGenomic DNA was isolated using the high salt method after peripheral blood collection in vacuum tubes.24 The selected probes corresponded to the following SNPs: FTO – rs9939609; AKTIP – rs9302648 and rs7189819; and AKT1 – rs2494738, rs3730358, and rs10149779. To improve statistical power, only SNPs with minor allele frequencies >0.2 in Caucasian and Yoruba individuals in the HapMap database were chosen. Genotyping was done with real-time polymerase chain reaction (RT-PCR) using a 7500 Real-Time PCR System (Applied Biosystems Inc., CA, USA), in allelic discrimination mode. PCR protocols were performed in accordance with the TaqMan® Genotyping Master Mix manufacturer's instructions (Applied Biosystems, CA, USA).
Statistical analysisAllele, haplotype and genotype frequencies were compared between groups with the chi-squared test using the UNPHASED software (UNPHASED, v.3.0.13, Cambridge, United Kingdom).25 1,000 permutations test (post-test) was performed for each test in order to estimate the global significance for all analyses and to validate the expectation-maximization values. HAPLOVIEW 4.1 software (HAPLOVIEW, v 4.1, Broad Institute, MA, USA) was used to evaluate pairwise linkage disequilibrium (LD) matrices between each SNP to examine the LD block structure and Hardy–Weinberg equilibrium (cutoff 0.05).26 Odds ratio and permutations were performed using UNPHASED. After Bonferroni correction, the significance level was established at 0.0083 for the genotype vs. group comparisons. Clinical characteristics of the obese/overweight group were compared with those of control group by one-way analysis of variance (ANOVA), followed by Tukey's post hoc test. Aiming to verify the detection of gene–gene interactions associated with complex human disease, multifactor dimensionality reduction (MDR) and Multifactor Dimensionality Reduction Pedigree Disequilibrium Test (MDR-PDT) were used.27
At least 10% of the samples were re-genotyped randomly for quality control, thus minimizing genotyping errors. An agreement of 100% between the results of both tests was achieved.
A secondary analysis tested if the SNPs included in the present study were linearly associated with HOMA-IR and BMI. A linear regression model was used to test the association of each independent predictor (SNP) with each metabolic outcome (HOMA-IR or BMI). Multiple linear regressions were used if more than one predictor showed association with each outcome.
ResultsA total of six SNPs were genotyped in a total of 348 individuals. All SNPs were in Hardy–Weinberg equilibrium and showed a minor allele frequency (MAF) higher than 10%, indicating a good penetration of the alleles in the population.
The clinical and laboratorial characteristics of the studied groups are presented in Table 1. As expected, the subjects in the obese/overweight group had higher BMI. In contrast to control group, the obese/overweight group showed higher fasting glucose, HOMA-IR index, total cholesterol, low-density lipoprotein, and triglycerides (p≤0.01; Table 1).
Demographic characteristics of study participants.
| Control n=153 (%) | Obese/overweight n=195 (%) | |
|---|---|---|
| Female | 91 (59.47) | 104 (53.33) |
| Age | 11.9 (±3.2) | 11.0 (±3.9) |
| BMI | 17.9 (±2.3) | 26.7 (±4.5) |
| SBP | 105.3 (±12.4) | 118.7 (±15.1) |
| DBP | 65.2 (±9.7) | 73.8 (±9.7) |
| Hypertension | 5 (3.2) | 61 (31.2) |
| Glucose (mg/dL) | 80.8 (±10.9) | 86.0 (±8.8) |
| HOMA-IR | 2.17 (±2.21) | 5.68 (±3.28) |
| Total cholesterol | 142 (±43) | 145 (±35) |
| LDL | 60.5 (±34) | 84.8 (±35) |
| HDL | 44 (±9) | 39 (±10) |
| Triglycerides (mg/dL) | 74.3 (±27) | 88.2 (±41) |
BMI, body mass index (kg/m2); SBP, systolic blood pressure (mmHg); DBP, diastolic blood pressure (mmHg); HOMA-IR, Homeostasis Model Assessment – Insulin Resistance; LDL, low-density lipoprotein (mg/dL); HDL, high-density lipoprotein (mg/dL).
Values are presented as mean±SD.
Gender and hypertension are samples numbers (%).
Table 2 shows the distribution of genotypes and alleles in the groups. In allelic analysis, the AKT1 rs10149779 showed a non-risk association with ancestral G allele (p=0.2; OR=0.70; χ2=4.40; df=1) between obese/overweight and control group after 1000 permutations test (post test). The same polymorphism also showed no genotypic association G/G (p=0.09; OR=0.61; χ2=6.29; df=2) in comparison with eutrophic subjects. The genotype T/T for marker rs9302648 AKTIP also did not show association (p=0.09; OR=1.03; χ2=7.15; df=2), after 1000 permutations test (post test). For other markers, no associations were observed (Table 2).
Genotype and allele frequencies of the six tagSNPs of FTO/AKT1/AKTIP polymorphisms in the sample.
| Obesity/overweight n=195 (%) | Control n=153 (%) | OR | 95% Lo 95% Hi | χ2 | df | p-value (1000 permut.) | |
|---|---|---|---|---|---|---|---|
| AKT1 | |||||||
| rs2494738 | |||||||
| G/G | 150 (76.92) | 126 (82.25) | 1 (ref) | 1.13 | 2 | 0.56 | |
| A/G | 30 (15.38) | 18 (11.76) | 1.40 | 0.73–2.63 | |||
| A/A | 01 (0.5) | 1 (0.65) | 0.84 | 0.05–13.57 | |||
| Ga | 330 (84.62) | 270 (88.24) | 1 (ref) | 0.83 | 1 | 0.36 | |
| A | 32 (8.2) | 20 (6.54) | 1.30 | 0.73–2.34 | |||
| rs3730358 | |||||||
| G/G | 116 (59.49) | 90 (58.82) | 1 (ref) | 2.31 | 2 | 0.31 | |
| A/G | 54 (27.69) | 48 (31.37) | 0.87 | 0.54–1.40 | |||
| A/A | 11 (5.64) | 4 (2.61) | 2.13 | 0.65–6.23 | |||
| Ga | 286 (73.33) | 228 (74.51) | 1 (ref) | 0.15 | 1 | 0.68 | |
| A | 76 (19.49) | 56 (18.3) | 1.08 | 0.73–1.59 | |||
| rs10149779 | |||||||
| G/G | 94 (48.21) | 55 (35.95) | 1 (ref) | 6.29 | 2 | 0.04 (0.09) | |
| G/A | 73 (37.44) | 76 (49.68) | 0.56 | 0.35–0.89 | |||
| A/A | 18 (9.23) | 17 (11.11) | 0.61 | 0.29–1.30 | |||
| Ga | 261 (37.83) | 186 (60.78) | 1 (ref) | 4.40 | 1 | 0.03 (0.20) | |
| A | 109 (27.95) | 110 (35.95) | 0.70 | 0.51–0.97 | |||
| AKTIP | |||||||
| rs9302648 | |||||||
| T/T | 76 (38.97) | 46 (30.07) | 1 (ref) | 7.15 | 2 | 0.02 (0.09) | |
| G/T | 76 (38.97) | 82 (53.59) | 0.56 | 0.34–0.90 | |||
| G/G | 36 (18.46) | 21 (13.73) | 1.03 | 0.54–1.98 | |||
| Ta | 228 (58.46) | 174 (56.86) | 1 (ref) | 0.34 | 1 | 0.55 | |
| G | 148 (37.95) | 124 (40.52) | 0.91 | 0.66–1.24 | |||
| rs7189819 | |||||||
| C/C | 67 (34.36) | 61 (39.87) | 1 (ref) | 0.95 | 2 | 0.61 | |
| C/T | 84 (43.08) | 65 (42.48) | 1.17 | 0.73–1.89 | |||
| T/T | 34 (17.44) | 23 (15.03) | 1.34 | 0.71–2.53 | |||
| Ca | 218 (55.9) | 187 (61.11) | 1 (ref) | 1.01 | 1 | 0.31 | |
| T | 152 (38.97) | 111 (36.27) | 1.17 | 0.85–1.60 | |||
| FTO | |||||||
| rs9939609 | rs9939609 | ||||||
| A/A | 44 (22.56) | 34 (22.22) | 1 (ref) | 4.42 | 2 | 0.10 | |
| T/A | 45 (23.08) | 71 (46.41) | 1.14 | 0.66–1.96 | |||
| T/T | 39 (20.0) | 46 (30.07) | 0.65 | 0.35–1.21 | |||
| Aa | 193 (49.49) | 139 (45.42) | 1 (ref) | 1.88 | 1 | 0.16 | |
| T | 183 (46.92) | 163 (53.27) | 0.80 | 0.59–1.09 | |||
Adjusted p-value from permutation test: 0.2 (allele); adjusted p-value from permutation test: 0.09 (genotypic); df, degree of freedom, df=1 between two alleles; df=2 between three genotypes; a significant association is indicated in bold and italic (<0.05); adjusted p-value from 1000 permutation test (multiple testing – Unphased).
OR, odds ratio.
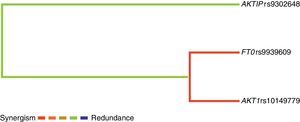
In the analysis of the interaction between genes through the multifactor dimensionality reduction, the dendogram indicated a relationship between the pairs of genes. The term synergy denotes a situation in which the interaction, based on the entropy between two SNPs, promotes more information than the correlation between the pair. In turn, redundancy is used in those cases in which the interaction amid two SNPs promotes less information than the correlation between the pairs (Fig. 1).
Dendrogram of gene–gene interaction of obese patients and controls. The colors of the caption comprise a spectrum representing the transition of synergism (black) for redundancy (gray). Observe the following interactions: synergism between rs9939609 FTO and rs10149779 AKT1 and redundancy between those and rs9302648 AKTIP.
No association was observed in the analysis of insulin resistance and those polymorphisms (data not shown).
Although a significant p-value for genetic analysis of FTO rs9939609 was not obtained at the gene–gene analysis, an interaction between this one and AKT1 rs10149779 was found.
Table 3 shows the result from linear regression models – no significant association was observed between each SNPs and HOMA-RI or BMI.
Linear regression model addressing the contributions of each SNP to HOMA-IR and BMI in the present sample.
| Linear regression models for HOMA | |||||
|---|---|---|---|---|---|
| Predictor | F | p | adjR2 | Std. beta | t |
| AKT1 rs2191738 HEX/FAM A/G | 0.00 | 0.983 | 0.00 | 0.01 | 0.22 |
| AKT1 rs3730358 HEX/FAM A/G | 0.12 | 0.725 | 0.00 | 0.02 | 0.35 |
| AKT1 rs10119779 HEX/FAM A/G | 0.02 | 0.880 | 0.00 | 0.01 | 0.15 |
| FTO rs9939309 HEX/FAM A/T | 0.00 | 0.971 | 0.00 | 0.00 | 0.03 |
| AKTIP rs9302318 HEX/FAM G/T | 0.65 | 0.421 | 0.00 | 0.05 | 0.81 |
| AKTIP rs7189819 HEX/FAM C/T | 0.33 | 0.567 | 0.00 | 0.04 | 0.57 |
| Linear regression models for BMI | |||||
|---|---|---|---|---|---|
| Predictor | F | p | adjR2 | Std. beta | t |
| AKT1 rs2191738 HEX/FAM A/G | 0.01 | 0.919 | 0.00 | 0.00 | 0.10 |
| AKT1 rs3730358 HEX/FAM A/G | 0.06 | 0.802 | 0.00 | −0.01 | −0.25 |
| AKT1 rs10119779 HEX/FAM A/G | 2.23 | 0.137 | 0.00 | −0.08 | −1.49 |
| FTO rs9939309 HEX/FAM A/T | 3.08 | 0.080 | 0.01 | −0.09 | 1.75 |
| AKTIP rs9302318 HEX/FAM G/T | 0.05 | 0.816 | 0.00 | 0.01 | 0.23 |
| AKTIP rs7189819 HEX/FAM C/T | 1.38 | 0.240 | 0.00 | −0.06 | −1.18 |
HOMA, homeostasis model assessment index; BMI, body mass index; adjR2, adjusted R2; std. beta, standardized regression coefficient.
Obesity is an intricate clinical condition modulated by many genetic and non-genetic factors, with several interactions among many of them. A total of six non-coding SNPs of AKT1, AKTIP, and FTO genes were investigated as candidate's genes for obesity in Brazilian children. Our results suggest that the AKT1, FTO, and AKTIP polymorphisms were not associated with obesity/overweight in Brazilian children. The fact that the obese/overweight children carrying the A allele of the FTO rs9939609 did not show an increased risk of becoming obese in this population is noteworthy. This polymorphism has been associated with BMI and type II diabetes in diverse populations, such as Finns and Asians.28–30 Recently, GWAS in Caucasians31 and Chinese children32 identified multiple single nucleotide polymorphisms (SNPs), including the FTO gene, as associated with BMI and obesity. Although several variants of the FTO gene have been associated to obesity in populations of European background, its effects in other ethnic populations remains to be established. In a study involving 478 African-American children, no association between variants of the FTO gene and BMI was found, a result similar to other studies with African-American and Gambian populations.33 The Brazilian ethnical blending might be one reason for the lack of association with a classical obesity-associated polymorphism.
Few studies regarding FTO variants and BMI in Latin American children and adolescents have been published to date. Da Silva et al.34 reported a significant association between the A/A genotype of the FTO rs9939609 and increased BMI and subcutaneous fat in 348 Brazilian children.35 Curiously, in another study regarding Brazilian children, the FTO rs9939609 A allele was associated with an increased BMI between vitamin D insufficient children, whereas no significant effect was observed among those with adequate vitamin D status. This might be a type I error, but it might also mean that this problem is more complex than expected, involving another metabolic factors.
To date, few studies have associated variants in the AKT1 gene and insulin resistance. In a recent study, a haplotype including the first exon of the AKT1 was associated with a higher frequency of metabolic syndrome in African-American and European-American subjects.36
In the present study, at the gene–gene analysis (MDR), an interaction between FTO rs9939609 and AKT1 rs10149779 was found. One possible reason for this difference between single polymorphism and MDR analysis results may be due to epistasis, defined as the interaction between two or more genes or their mRNA protein products modulating a phenotype.37 Statistical analyzes and animal models have shown that epistasis is a common phenomenon in obesity. Thus, genes for obesity confer, in isolation, small increases in the risk of obesity. However, they may influence the development of obesity by more complex mechanisms, especially gene vs. gene and gene-environment interactions.38
As some studies indicated that FTO had larger effects on childhood BMI at higher percentiles,39 a linear regression was performed; however, no such association was found. Some negative association findings might result from some limitations of the study, mainly the small sample size. Moreover, the study did not consider environmental factors, such as the patterns of physical activity, the presence of overweight/obese parents, and the data concerning food intake, which prevents further analysis.
Obesity is complex disease and the mechanisms involved in the interaction between genotype and environment still need to be clarified; the present data may shed some light to better understand such a complex and relevant health problem. It should be taken in account the existing publication bias in studies with candidate gene approach. Furthermore, many studies on obesity genetics can present an inadequate statistical power, thereby increasing the risk of false positives associations.40
In summary, no evidence of association between AKT1, AKTIP and FTO polymorphisms with obesity and insulin resistance was observed in this sample of Brazilian children. There are several reasons for the contrasting results observed within the studies, including differences in the genetic background of the different samples. Considering the associations found in other studies, the authors believe that studies involving a larger number of individuals with different ethnic groups may be required to elucidate the role of these genes with obesity. Thus, the present study emphasizes the importance of caution when analyzing candidate-gene studies and reinforces the need for well-conducted studies to expand the understanding of the genetic and environmental factors involved in obesity and insulin resistance.
FundingCAPES, FAPESP, CNPq (FAPEMIG: CBB-APQ-00075-09/CNPq 573646/2008-2), and FAPEMIG: CDS-BPD-00296-14.
Conflicts of interestThe authors declare no conflicts of interest.
This study was supported by CAPES, FAPESP, CNPq (FAPEMIG: CBB-APQ-00075-09/CNPq 573646/2008-2), and FAPEMIG: CDS-BPD-00296-14.
Both Authors contributed equally to this paper.
Please cite this article as: Pereira PA, Alvim-Soares AM, Sandrim VC, Lanna CM, Souza-Costa DC, Belo VA, et al. Lack of association between genetic polymorphism of FTO, AKT1 and AKTIP in childhood overweight and obesity. J Pediatr (Rio J). 2016;92:521–7.