The prevalence of non-alcoholic fatty liver disease in children has risen significantly, owing to the worldwide childhood obesity epidemic in the last two decades. Non-alcoholic fatty liver disease is closely linked to sedentary lifestyle, increased body mass index, and visceral adiposity. In addition, individual genetic variations also have a role in the development and progression of non-alcoholic fatty liver disease. The aim of this study was to investigate the gene polymorphisms of MCP-1 (-2518 A/G) (rs1024611), CCR-2 (190 G/A) (rs1799864), ABCA1 (883 G/A) (rs4149313), and IL-17A (-197 G/A) (rs2275913) in obese Turkish children with non-alcoholic fatty liver disease.
MethodsThe study recruited 186 obese children aged 10–17 years, including 101 children with non-alcoholic fatty liver disease and 85 children without non-alcoholic fatty liver disease. Anthropometric measurements, insulin resistance, a liver panel, a lipid profile, liver ultrasound examination, and genotyping of the four variants were performed.
ResultsNo difference was found between the groups in respect to age and gender, body mass index, waist/hip ratio, or body fat ratio. In addition to the elevated ALT levels, AST and GGT levels were found significantly higher in the non-alcoholic fatty liver disease group compared to the non non-alcoholic fatty liver disease group (p<0.05). The A-allele of IL-17A (-197 G/A) (rs2275913) was associated with non-alcoholic fatty liver disease (odds ratio [OR] 2.05, 95% confidence interval: 1.12–3.77, p=0.02).
ConclusionsThe findings of this study suggest that there may be an association between IL-17A (-197 G/A) (rs2275913) polymorphism and non-alcoholic fatty liver disease development in obese Turkish children.
A prevalência de doença hepática gordurosa não alcoólica em crianças aumentou significativamente devido à epidemia de obesidade infantil em todo o mundo nas últimas duas décadas. A doença hepática gordurosa não alcoólica está intimamente ligada ao estilo de vida sedentário, ao aumento do índice de massa corporal e à adiposidade visceral. Além disso, variações genéticas individuais também têm um papel no desenvolvimento e na progressão da doença hepática gordurosa não alcoólica. O objetivo deste estudo foi investigar os polimorfismos genéticos MCP-1 (-2518 A/G) (rs1024611), CCR-2 (190 G/A) (rs1799864), ABCA1 (883 G/A) (rs4149313) e IL-17A (-197 G/A) (rs2275913) em crianças turcas obesas com doença hepática gordurosa não alcoólica.
MétodosO estudo recrutou 186 crianças obesas entre 10 e 17 anos, inclusive 101 crianças com doença hepática gordurosa não alcoólica e 85 crianças sem doença hepática gordurosa não alcoólica. Medidas antropométricas, resistência à insulina, painel hepático, perfil lipídico, exame ultrassonográfico do fígado e genotipagem de quatro variantes foram feitos.
ResultadosNenhuma diferença foi encontrada entre os grupos em relação à idade e sexo, índice de massa corporal, relação cintura/quadril ou proporção de gordura corporal. Além dos níveis elevados de ALT, os níveis de AST e GGT foram significativamente maiores no grupo doença hepática gordurosa não alcoólica em comparação com o grupo não doença hepática gordurosa não alcoólica (p<0,05). O alelo A de IL-17A (-197 G/A) (rs2275913) foi associado à doença hepática gordurosa não alcoólica (odds ratio [OR] 2,05, intervalo de confiança de 95%: 1,12-3,77, p=0,02).
ConclusõesOs achados deste estudo sugerem que pode haver uma associação entre o polimorfismo IL-17A (-197 G/A) (rs2275913) e o desenvolvimento da doença hepática gordurosa não alcoólica em crianças turcas obesas.
Non-alcoholic fatty liver disease (NAFLD) is a chronic liver disease caused by excessive fat accumulation in the liver, without a history of chronic alcohol consumption, metabolic liver diseases (Wilson disease), or congenital, viral, or autoimmune liver diseases.1 NAFLD is included in a broad spectrum of liver damage, ranging from simple steatosis to steatohepatitis, fibrosis, and even cirrhosis.1 The prevalence of NAFLD has increased significantly due to the worldwide childhood obesity epidemic in the last two decades.2 NAFLD is closely linked to a sedentary lifestyle, increased body mass index (BMI), and visceral adiposity in children, resulting from excess caloric intake.3 Several recent studies have reported that, in addition to environmental risk factors, individual genetic variations may also contribute to the development and progression of NAFLD.4
Several candidate genes have been implicated in the pathogenesis of NAFLD, including genes involved in hepatic lipid metabolism, insulin sensitivity, and generation of reactive oxidant species or cytokines.5 Single-nucleotide polymorphisms in genes involved in lipid metabolism (patatin-like phospholipase domain-containing 3 and lipin 1), oxidative stress (superoxide dismutase 2), insulin signalling (insulin receptor substrate-1), and fibrogenesis (Krüppel-like factor 6) have been found to be associated with NAFLD in children.2 Apart from these well-known polymorphisms, polymorphisms in genes that encode proteins known to be involved in the pathogenesis of NAFLD may also be associated with NAFLD development.
The monocyte chemo-attractant protein-1 (MCP-1), also called chemokine ligand 2 (CCL2), a strong chemotactic factor, plays a role in the activation of monocytes, macrophages, and T-cells in the acute and chronic phases of inflammation.6 The 2518 A/G polymorphism in the MCP-1 gene affects the level of MCP-1 expression in response to inflammatory stimuli, and is known to be associated with diabetes mellitus and several auto-immune diseases.7,8 MCP-1 exerts a chemotactic effect on monocytes and T-cells via the chemokine receptor 2 (CCR2).9 The 190 G/A polymorphism in the CCR gene has also been reported to be associated with NAFLD development.10
ATP-binding cassette transporter 1 (ABCA1) is a member of the ATP-binding cassette (ABC) membrane transporter family. ABCA1 expression resulted in increased cholesterol efflux and decreased hepatic lipid contents.11 Inflammatory stress increases cholesterol accumulation in hepatocytes and inhibits the expression of ABCA1.12 Furthermore, it has been reported that decreased hepatic ABCA1 expression can cause steatohepatitis in morbidly obese adult patients.11 Interleukin-17 (IL-17) is a pro-inflammatory molecule produced by T-helper 17 cells (Th17). It mediates the immune response against extra-cellular bacterial and fungal pathogens, and is involved in the development of inflammatory and auto-immune diseases.13 IL-17 has also been suggested to play a critical role in the development of various liver diseases, such as chronic viral hepatitis, auto-immune diseases, and alcoholic liver diseases.14–16 IL-17A (-197 G/A) has been shown to affect the development of ulcerative colitis and rheumatoid arthritis.17
The present study aimed to investigate the association between NAFLD and polymorphisms in genes encoding proteins that have been suggested to play a role in NAFLD pathogenesis. To the best of the authors’ knowledge, the relationship between the four single-nucleotide polymorphisms and NAFLD has not been studied previously.
MethodsThe study included obese Turkish patients aged 10–17 years, who presented at the Paediatric Gastroenterology, Hepatology, and Nutrition Clinic and the Paediatric Endocrinology Clinic of the Kanuni Training and Research Hospital between June, 2015 and July, 2016. Patients were considered obese if they had BMI Z-scores ≥2 for age and gender.18 The 186 patients enrolled in the study were separated into two groups. The NAFLD group (n=101) comprised patients diagnosed with NAFLD, with alanine aminotransferase (ALT) levels greater than twice the upper limit of the normal level (males>50U/L, females>44U/L) and fatty liver disease detected by ultrasonography (USG).19 Patients with other causes of fatty liver disease, such as Wilson disease, α-1 antitrypsin deficiency, auto-immune hepatitis, and viral hepatitis, were excluded. The non-NAFLD group (n=85) included obese patients without NAFLD (normal ALT levels and USG imaging of liver), having similar anthropometric measurements, insulin resistance, and lipid profile as the NAFLD group. None of the patients in the study underwent liver biopsy. Patients with genetic, endocrine, or metabolic diseases that might cause obesity were not included in the study. In addition, none of the patients had any co-morbid proinflammatory diseases such as inflammatory bowel disease.
The study was performed after receiving approval from the local ethics committee (Registry Url: 2015/27; Identifier: Trabzon Kanuni Training and Research Hospital Clinical Research Ethics Committee) and informed parental consent, in accordance with the Declaration of Helsinki.
All participants were subjected to physical examination. Height was measured to the nearest centimetre using a Harpenden stadiometer (Holtain Limited – Crymych, Dyfed, United Kingdom). Weight was measured unclothed to the nearest 0.1kg using a calibrated balance scale. Body mass index (BMI) was calculated using the formula: weight (kg)/height (m2). Z-scores for BMI were calculated using the reference values for Turkish children.20 Body fat ratio was calculated with the bioelectrical impedance analysis (BIA) method on a Tanita BC 418 (Tanita Corporation – Tokyo, Japan) device. Waist and hip circumference measurements were obtained using a non-stretch tape measure while the patient stood upright with the feet close together (12–15cm) and arms by the side. The waist-hip ratio was calculated by dividing the waist measurement by the hip measurement.
After a ten-hour fast, peripheral venous blood samples were obtained to determine the levels of insulin (Beckman Coulter DXI 800 – California, United States), high-density lipoprotein (HDL), low-density lipoprotein (LDL), triglycerides, total cholesterol, ALT, aspartate aminotransferase (AST), gamma glutamyltransferase (GGT), and glucose (Beckman Coulter AU5821 – California, United States). The homeostatic model assessment of insulin resistance (HOMA-IR) value was calculated using the formula: glucose×insulin (μU/mL)/405.21
All examinations were performed by two radiologists with more than ten years of experience in paediatric abdominal imaging and liver steatosis, and the results were determined by consensus. The radiologists were blinded to the clinical findings and laboratory results of the patients. The ultrasound machine used was an Aplio 500 (Toshiba Medical Systems – Otawara, Japan) with linear 4–9MHz transducers. The patients were evaluated in the dorsal decubitus position with the right arm at maximum abduction. The patients were asked to fast for four hours prior to the examination. Longitudinal images of the right liver lobe and the ipsilateral kidney were obtained, including the sagittal hepatic sections. Semi-quantitative grading of fatty changes was performed using the USG findings of fatty liver, vascular blurring, and deep attenuation with liver-kidney contrast.22
DNA was isolated from the peripheral blood using the automatic QuickGene device. Target areas were amplified using PCR with primary pairs: for MCP1 (2518 A/G) polymorphism, F: 5′ CTTTCCCTTGTGTGTCCCC 3′, R: 5′ TTACTCCTTTTCTCCCCAACC 3′; for CCR-2 rs1799864 polymorphism, F: 5′ ATTTCCCCAGTACATCCACAAC 3′, R: 5′ CCCACAATGGGAGAGTAATAAG 3′; for ABCA1rs4149313 polymorphism, F: 5′ GAGAAGAGCCACCCTGGTTCCAACCAGAAGAGGAT 3′, R: 5′ AGAAAGGCAGGAGACAT CGCTT 3′; and for IL-17A rs2275913 polymorphism, F: 5′ AACAAGTAAGAATGAAAAGAGGACATGGT 3′, R: 5′ CCCCCAATGAGGTCATAGAAGAATC 3′.
Sectioning was performed using Pvull (Vivantis) restriction enzymes for MCP1 (-2518 A/G) genotype assignment, FokL (Vivantis) restriction enzymes for CCR-2 rs1799864 genotype assignment, Econl (Vivantis) restriction enzymes for ABCA1 rs4149313 genotype assignment, and BstENI restriction enzymes for IL-17A rs2275913 genotype assignment. After cutting by 2% agarose gel electrophoresis, analysis was performed according to the product size. For control, accuracy was confirmed by 10% random sequence analysis from both groups.
The data were evaluated using SPSS 13.0 (SPSS Inc. – Chicago, IL, United States) statistics software. Descriptive data were presented as mean±standard deviation (SD). The analysis of the results was performed using percentage distribution for qualitative data and median interquartile range (IQR) or mean (standard deviation) for quantitative data. For comparison between two groups, Student's t-test for independent paired samples was used for variables showing normal distribution and the Mann–Whitney U-test was used for those not distributed normally. In case of more than two groups, the ANOVA test was used for variables showing normal distribution and the Kruskal–Wallis test was used for those not showing normal distribution. The chi-squared test was used for comparing categorical variables. Genotypic association and the odds ratio (OR) with 95% confidence interval (CI) were estimated by binary logistic regression analysis. In all cases, p-values less than 0.05 were considered statistically significant. Power analysis was performed using normal approximation with the continuity correction method of Open Epi (3.01 version).
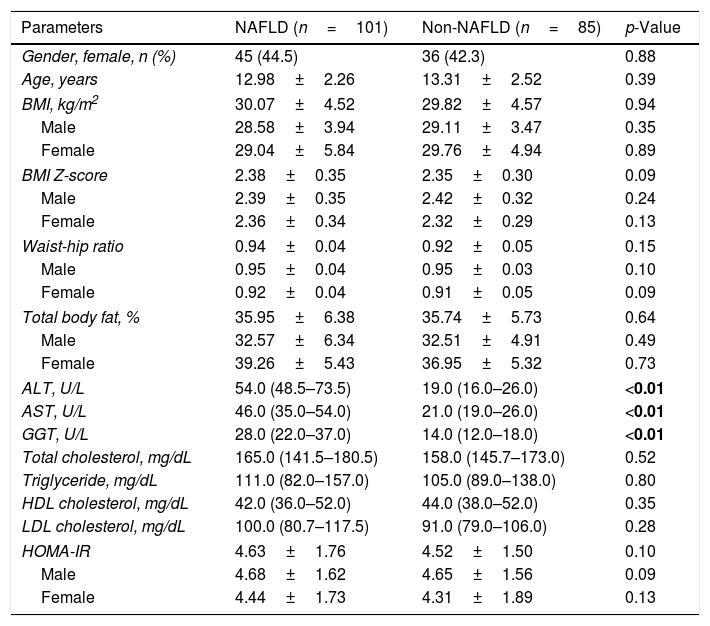
ResultsThe demographic characteristics, anthropometric measurements, and laboratory data of the patients are shown in Table 1. No differences were found between the groups in terms of age and gender, BMI, waist/hip ratio, or body fat ratio. In addition to ALT, AST and GGT concentrations were also found to be significantly higher in the NAFLD group than the non-NAFLD group (p<0.05).
Characteristics of the groups.
| Parameters | NAFLD (n=101) | Non-NAFLD (n=85) | p-Value |
|---|---|---|---|
| Gender, female, n (%) | 45 (44.5) | 36 (42.3) | 0.88 |
| Age, years | 12.98±2.26 | 13.31±2.52 | 0.39 |
| BMI, kg/m2 | 30.07±4.52 | 29.82±4.57 | 0.94 |
| Male | 28.58±3.94 | 29.11±3.47 | 0.35 |
| Female | 29.04±5.84 | 29.76±4.94 | 0.89 |
| BMI Z-score | 2.38±0.35 | 2.35±0.30 | 0.09 |
| Male | 2.39±0.35 | 2.42±0.32 | 0.24 |
| Female | 2.36±0.34 | 2.32±0.29 | 0.13 |
| Waist-hip ratio | 0.94±0.04 | 0.92±0.05 | 0.15 |
| Male | 0.95±0.04 | 0.95±0.03 | 0.10 |
| Female | 0.92±0.04 | 0.91±0.05 | 0.09 |
| Total body fat, % | 35.95±6.38 | 35.74±5.73 | 0.64 |
| Male | 32.57±6.34 | 32.51±4.91 | 0.49 |
| Female | 39.26±5.43 | 36.95±5.32 | 0.73 |
| ALT, U/L | 54.0 (48.5–73.5) | 19.0 (16.0–26.0) | <0.01 |
| AST, U/L | 46.0 (35.0–54.0) | 21.0 (19.0–26.0) | <0.01 |
| GGT, U/L | 28.0 (22.0–37.0) | 14.0 (12.0–18.0) | <0.01 |
| Total cholesterol, mg/dL | 165.0 (141.5–180.5) | 158.0 (145.7–173.0) | 0.52 |
| Triglyceride, mg/dL | 111.0 (82.0–157.0) | 105.0 (89.0–138.0) | 0.80 |
| HDL cholesterol, mg/dL | 42.0 (36.0–52.0) | 44.0 (38.0–52.0) | 0.35 |
| LDL cholesterol, mg/dL | 100.0 (80.7–117.5) | 91.0 (79.0–106.0) | 0.28 |
| HOMA-IR | 4.63±1.76 | 4.52±1.50 | 0.10 |
| Male | 4.68±1.62 | 4.65±1.56 | 0.09 |
| Female | 4.44±1.73 | 4.31±1.89 | 0.13 |
BMI, body mass index; ALT, alanine aminotransferase; AST, aspartate aminotransferase; GGT, gamma glutamyltransferase; HOMA-IR, homeostesis model assessment-estimated insulin resistance; NAFLD, nonalcoholic fatty liver disease; HDL, high-density lipoprotein; LDL, low-density lipoprotein.
Values are expressed as median (25th to 75th percentiles) or mean±SD, as appropriate.
Bold value indicates a p-value less than 0.05.
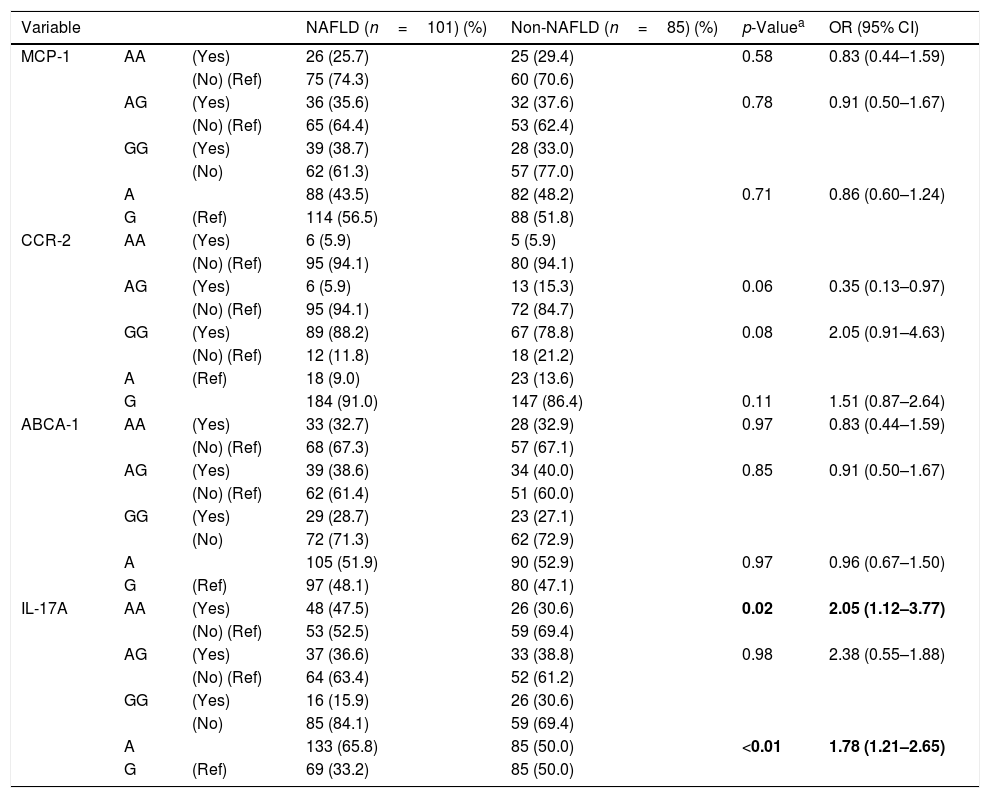
No differences were observed between the groups in terms of genotype and allele frequency for MCP-1 rs1024611, CCR-2 rs1799864, and ABCA1 rs4149313 polymorphisms (Table 2). Among the four variants, only IL-17A rs2275913 was found to be associated with NAFLD. The percentage of IL-17A rs2275913 AA genotypes was significantly higher in the NAFLD group than the non-NAFLD group (p=0.02, OR: 2.05, 95% CI: 1.12–3.77). The frequency of the A allele of IL-17A rs2275913 was significantly higher in the NAFLD group than the non-NAFLD group (p=<0.01). The power of the study was calculated as 53%. For this power (alpha error: 0.05 and Df: 1), the effect size was calculated as 0.15.
Genotypic and allelic frequency of the MCP-1 rs1024611, CCR-2 rs1799864, ABCA1 rs4149313, and IL-17A rs2275913 polymorphisms in patients, and logistic regression analysis of predictive factors for NAFLD.
| Variable | NAFLD (n=101) (%) | Non-NAFLD (n=85) (%) | p-Valuea | OR (95% CI) | ||
|---|---|---|---|---|---|---|
| MCP-1 | AA | (Yes) | 26 (25.7) | 25 (29.4) | 0.58 | 0.83 (0.44–1.59) |
| (No) (Ref) | 75 (74.3) | 60 (70.6) | ||||
| AG | (Yes) | 36 (35.6) | 32 (37.6) | 0.78 | 0.91 (0.50–1.67) | |
| (No) (Ref) | 65 (64.4) | 53 (62.4) | ||||
| GG | (Yes) | 39 (38.7) | 28 (33.0) | |||
| (No) | 62 (61.3) | 57 (77.0) | ||||
| A | 88 (43.5) | 82 (48.2) | 0.71 | 0.86 (0.60–1.24) | ||
| G | (Ref) | 114 (56.5) | 88 (51.8) | |||
| CCR-2 | AA | (Yes) | 6 (5.9) | 5 (5.9) | ||
| (No) (Ref) | 95 (94.1) | 80 (94.1) | ||||
| AG | (Yes) | 6 (5.9) | 13 (15.3) | 0.06 | 0.35 (0.13–0.97) | |
| (No) (Ref) | 95 (94.1) | 72 (84.7) | ||||
| GG | (Yes) | 89 (88.2) | 67 (78.8) | 0.08 | 2.05 (0.91–4.63) | |
| (No) (Ref) | 12 (11.8) | 18 (21.2) | ||||
| A | (Ref) | 18 (9.0) | 23 (13.6) | |||
| G | 184 (91.0) | 147 (86.4) | 0.11 | 1.51 (0.87–2.64) | ||
| ABCA-1 | AA | (Yes) | 33 (32.7) | 28 (32.9) | 0.97 | 0.83 (0.44–1.59) |
| (No) (Ref) | 68 (67.3) | 57 (67.1) | ||||
| AG | (Yes) | 39 (38.6) | 34 (40.0) | 0.85 | 0.91 (0.50–1.67) | |
| (No) (Ref) | 62 (61.4) | 51 (60.0) | ||||
| GG | (Yes) | 29 (28.7) | 23 (27.1) | |||
| (No) | 72 (71.3) | 62 (72.9) | ||||
| A | 105 (51.9) | 90 (52.9) | 0.97 | 0.96 (0.67–1.50) | ||
| G | (Ref) | 97 (48.1) | 80 (47.1) | |||
| IL-17A | AA | (Yes) | 48 (47.5) | 26 (30.6) | 0.02 | 2.05 (1.12–3.77) |
| (No) (Ref) | 53 (52.5) | 59 (69.4) | ||||
| AG | (Yes) | 37 (36.6) | 33 (38.8) | 0.98 | 2.38 (0.55–1.88) | |
| (No) (Ref) | 64 (63.4) | 52 (61.2) | ||||
| GG | (Yes) | 16 (15.9) | 26 (30.6) | |||
| (No) | 85 (84.1) | 59 (69.4) | ||||
| A | 133 (65.8) | 85 (50.0) | <0.01 | 1.78 (1.21–2.65) | ||
| G | (Ref) | 69 (33.2) | 85 (50.0) | |||
NAFLD, nonalcoholic fatty liver disease; OR, odds ratio; CI, confidence interval; Ref, reference.
Bold value indicates a p-value less than 0.05.
As compared with the reference genotype group, the differences in genotype frequencies were analyzed by using the chi-squared test. p<0.05 was considered to be statistically significant. All four polymorphisms were included in multivariate log regression analysis at the same time.
Models: MCP-1 AA, AG genotype and A allele; CCR-2AG, GG genotype and G allele; ABCA-1 AA, AG genotype and A allele; IL-17A AA, AG genotype and A allele. Dependent variable: presence of NAFLD.
Univariate logistic regression analysis showed that patients with the IL-17A rs2275913 AA genotype had a 3.00 (95% CI: 1.37–6.58; p≤0.01) times greater chance of having NAFLD than patients with the GG genotype (Table 3).
Univariate logistic regression analysis of predictive factors for NAFLD.
| Variable | Coefficient estimate | SE | OR (95% CI) | p-Value |
|---|---|---|---|---|
| IL-17A rs2275913 AA genotypea | 1.10 | 0.40 | 3.00 (1.37–6.58) | <0.01 |
| IL-17A rs2275913 AG genotype | 0.60 | 0.40 | 1.82 (0.83–3.97) | 0.13 |
NAFLD, nonalcoholic fatty liver disease; OR, odds ratio; CI, confidence interval.
Bold value indicates a p-value less than 0.05.
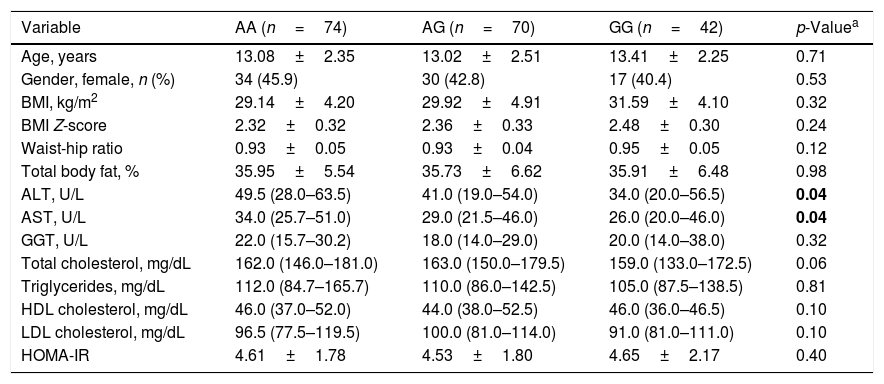
The clinical and biochemical characteristics of the groups, with respect to the IL-17A rs2275913 genotype, are shown in Table 4. There were significant differences in AST and ALT concentrations between the three groups (p=0.04 and p=0.04, respectively). The serum ALT and AST concentrations were highest in the AA genotype and lowest in the GG genotype.
Basic characteristics of the subjects across IL-17A rs2275913 genotypes.
| Variable | AA (n=74) | AG (n=70) | GG (n=42) | p-Valuea |
|---|---|---|---|---|
| Age, years | 13.08±2.35 | 13.02±2.51 | 13.41±2.25 | 0.71 |
| Gender, female, n (%) | 34 (45.9) | 30 (42.8) | 17 (40.4) | 0.53 |
| BMI, kg/m2 | 29.14±4.20 | 29.92±4.91 | 31.59±4.10 | 0.32 |
| BMI Z-score | 2.32±0.32 | 2.36±0.33 | 2.48±0.30 | 0.24 |
| Waist-hip ratio | 0.93±0.05 | 0.93±0.04 | 0.95±0.05 | 0.12 |
| Total body fat, % | 35.95±5.54 | 35.73±6.62 | 35.91±6.48 | 0.98 |
| ALT, U/L | 49.5 (28.0–63.5) | 41.0 (19.0–54.0) | 34.0 (20.0–56.5) | 0.04 |
| AST, U/L | 34.0 (25.7–51.0) | 29.0 (21.5–46.0) | 26.0 (20.0–46.0) | 0.04 |
| GGT, U/L | 22.0 (15.7–30.2) | 18.0 (14.0–29.0) | 20.0 (14.0–38.0) | 0.32 |
| Total cholesterol, mg/dL | 162.0 (146.0–181.0) | 163.0 (150.0–179.5) | 159.0 (133.0–172.5) | 0.06 |
| Triglycerides, mg/dL | 112.0 (84.7–165.7) | 110.0 (86.0–142.5) | 105.0 (87.5–138.5) | 0.81 |
| HDL cholesterol, mg/dL | 46.0 (37.0–52.0) | 44.0 (38.0–52.5) | 46.0 (36.0–46.5) | 0.10 |
| LDL cholesterol, mg/dL | 96.5 (77.5–119.5) | 100.0 (81.0–114.0) | 91.0 (81.0–111.0) | 0.10 |
| HOMA-IR | 4.61±1.78 | 4.53±1.80 | 4.65±2.17 | 0.40 |
BMI, body mass index; ALT, alanine aminotransferase; AST, aspartate aminotransferase; GGT, gamma glutamyltransferase; HOMA-IR, homeostesis model assessment-estimated insulin resistance; HDL, high-density lipoprotein; LDL, low-density lipoprotein.
Bold value indicates a p-value less than 0.05.
This study evaluated the effect of four polymorphisms in the development of NAFLD in obese children. The AA genotype of the IL-17A gene was found more frequently in obese children with NAFLD than in obese children without NAFLD. However, no significant differences were found in MCP-1 rs1024611, CCR-2 rs1799864, and ABCA1 rs4149313 gene polymorphisms between obese Turkish children with and without NAFLD, suggesting that these polymorphisms had no role in the development of NAFLD in obese children.
The balance between pro-inflammatory and anti-inflammatory mechanisms is of critical importance for the development and progression of NAFLD. Stimulation of pro-inflammatory macrophages by pro-inflammatory mediators such as interferon-γ and lipopolysaccharides stimulates the secretion of pro-inflammatory cytokines such as TNFα, IL-6, IL-17, and IL-23, which in turn causes hepatic and metabolic damages.23,24 In contrast, regulatory T-cells (Treg) prevent the production of pro-inflammatory cytokines such as IL-17 by expressing anti-inflammatory cytokines such as IL-10, thus controlling inflammation.25 In experimental studies, the activation of the IL-17 pathway has been shown to play a significant role in the development of NAFLD, progression of steatosis, and formation of fibrosis.26
The IL-17A rs2275913 polymorphism is known to be strongly associated with IL-17 secretion from T-cells. In an in vitro study by Espinoza et al., a higher level of IL-17 secretion was observed in stimulated T-cells of subjects with the IL-17A rs2275913 AA allele than in subjects without it.27 Differential binding of the allelic variants of the IL-17A rs2275913 polymorphism to the nuclear factor activated T-cells (NFAT) was proposed to be responsible for the differences in IL-17A secretion.27 Several inflammatory diseases have been found to be clinically associated with the IL-17A rs2275913 polymorphism. The rs2275913 AA homozygote was found to be associated with increased risk of susceptibility to rheumatoid arthritis in Caucasian populations and ulcerative colitis in Japanese populations.28,29
In the present study, it was found that the IL-17A rs2275913 polymorphism was more frequent in obese children with NAFLD than in those without NAFLD. A previous study has also reported an association between IL-17 concentration and ALT concentration in patients with chronic hepatitis B virus infection, reflecting the degree of liver damage.30 Consistent with these observations, serum ALT as well as serum AST concentrations significantly differed among the IL-17 rs2275913 allele groups (AA, AG, and GG). Serum ALT and AST concentrations were highest in the AA genotype and lowest in the GG genotype. These findings suggested that the AA genotype could be a risk factor for the development of NAFLD in obese patients.
No significant differences were found in MCP-1 rs1024611, CCR-2 rs1799864, and ABCA1 rs4149313 gene polymorphisms between obese children with and without NAFLD, indicating that these polymorphisms did not appear to play a role in the development of NAFLD. However, these polymorphisms may well be related to the development of steatohepatitis in obese children, as it was not possible to assess the degree of hepatic fibrosis and inflammation in these patients.
This study has some limitations. First, the study population was relatively small. The frequency of the studied polymorphisms is known to vary among races. The ethnic structure of the population could affect the prevalence of the polymorphisms. Therefore, these results cannot be generalized to other populations. Second, NAFLD was diagnosed by ultrasonography and by measuring elevated transaminases levels, which are regarded as imperfect tools for detecting steatosis. None of the patients in this study underwent liver biopsy or fibroscan. Therefore, neither fibrosis nor liver inflammation could be assessed in the participants. Consequently, the association between the polymorphisms and the degree of fibrosis and inflammation could not be examined. Third, the effect of the IL-17A rs2275913 polymorphism on gene transcription was not investigated.
In conclusion, the results of this study showed that the AA genotype of the IL-17A gene may be associated with NAFLD development. This polymorphism may serve as a predictor for liver steatosis and provide useful insights for identifying potential therapeutic targets for treating liver diseases in obese patients. Unlike IL-17A (-197 G/A) (rs2275913), no relation was found between the MCP-1 (-2518 A/G) (rs1024611), CCR-2 (190 G/A) (rs1799864), and ABCA1 (883 G/A) (rs4149313) polymorphisms and NAFLD in obese Turkish children. Further large-scale studies on the association between these polymorphisms and NAFLD, as well as fibrosis, need to be performed in different ethnicities in order to confirm these findings.
FundingThis work was supported by the Kanuni Training and Research Hospital.
Conflicts of interestThe authors declare no conflicts of interest.
Ethics committee approvalEthics committee approval was received for this study from the ethics committee of Kanuni Training and Research Hospital.
Please cite this article as: Akbulut UE, Emeksiz HC, Citli S, Cebi AH, Korkmaz HA, Baki G. IL-17A, MCP-1, CCR-2, and ABCA1 polymorphisms in children with non-alcoholic fatty liver disease. J Pediatr (Rio J). 2019;95:350–7.