Respiratory syncytial virus is a pathogen frequently involved in nosocomial outbreaks. Although several studies have reported nosocomial outbreaks in neonatal intensive care units, molecular epidemiology data are scarce. Here, the authors describe two consecutive respiratory syncytial virus outbreaks caused by genotypes ON-1 and NA-2 in a neonatal intensive care unit in São Paulo, Brazil.
MethodsA prospective search for respiratory syncytial virus was performed after diagnosing the index case and four other symptomatic newborns in the neonatal intensive care unit. Nasopharyngeal aspirate samples of all patients in the neonatal intensive care unit were tested for 17 respiratory viruses using real-time reverse transcriptase polymerase chain reaction. Genotyping was performed using nucleotide sequencing.
ResultsFrom May to August 2013, two different outbreaks were detected in the neonatal intensive care unit. A total of 20 infants were infected with respiratory syncytial virus-A (ten and 14 with ON-1 and NA-2 genotypes, respectively). The mean age of the infants was 10 days, mean birth weight was 1,961g, and the mean gestational age was 33 weeks. Risk factors (heart disease, lung disease, and prematurity) were present in 80% and 85.7% of infants in the ON-1 and NA-2 groups, respectively. In total, 45.8% of infants were asymptomatic and 20.8% required mechanical ventilation. Coinfections were not detected during the outbreaks.
ConclusionsInfants in a neonatal intensive care unit who develop abrupt respiratory symptoms should be tested for respiratory viruses, especially respiratory syncytial virus. Even in the absence of severe symptoms, respiratory syncytial virus detection can prevent nosocomial transmission through infection control measures. A better understanding of respiratory syncytial virus molecular epidemiology is essential for developing new vaccines and antiviral drugs against respiratory syncytial virus.
O vírus sincicial respiratório é um patógeno frequentemente envolvido em surtos nosocomiais. Embora vários estudos tenham relatado tais surtos em unidades de terapia intensiva neonatal, os dados epidemiológicos moleculares são escassos. Neste artigo, descrevemos dois surtos consecutivos de vírus sincicial respiratório causados pelos genótipos ON-1 e NA-2 em uma unidade de terapia intensiva neonatal em São Paulo, Brasil.
MétodosUma busca prospectiva por vírus sincicial respiratório foi realizada após o diagnóstico do caso índice e outros quatro recém-nascidos sintomáticos na unidade de terapia intensiva neonatal. Amostras de aspirado nasofaríngeo de todos os pacientes da unidade de terapia intensiva neonatal foram testadas para 17 vírus respiratórios com reação em cadeia da polimerase via transcriptase reversa em tempo real. A genotipagem foi realizada utilizando sequenciamento de nucleotídeos.
ResultadosDe maio a agosto de 2013, foram detectados dois surtos diferentes na unidade de terapia intensiva neonatal. Vinte e quatro crianças foram infectadas com vírus sincicial respiratório-A (10 e 14 com os genótipos ON-1 e NA-2, respectivamente). A média da idade dos lactentes era de 10 dias, o peso médio ao nascer foi de 1961g e a idade gestacional média de 33 semanas. Fatores de risco (doença cardíaca, doença pulmonar e prematuridade) estavam presentes em 80% e 85,7% dos bebês nos grupos ON-1 e NA-2, respectivamente. No total, 45,8% dos lactentes eram assintomáticos e 20,8% necessitaram de ventilação mecânica. Não foram detectadas coinfecções durante os surtos.
ConclusõesBebês em unidade de terapia intensiva neonatal que desenvolvem sintomas respiratórios abruptos devem ser testados para vírus respiratórios, especialmente o vírus sincicial respiratório. Mesmo na ausência de sintomas graves, a detecção de vírus sincicial respiratório pode prevenir a transmissão nosocomial através de medidas de controle de infecção. Um melhor entendimento da epidemiologia molecular do vírus sincicial respiratório é essencial para o desenvolvimento de novas vacinas e drogas antivirais contra o vírus sincicial respiratório.
Respiratory syncytial virus (RSV) infection is potentially life-threatening in infants, particularly in premature infants and those with chronic lung disease or congenital heart disease. RSV causes severe outbreaks in neonatal intensive care units (NICUs), leading to substantial morbidity and mortality in preterm infants, as well as increased financial costs.1,2 Conventional infection control methods, such as hand hygiene and patient isolation in cohorts, are recommended; however, these procedures may be supplemented by the use of palivizumab (Synagis® – MedImmune Inc., Gaithersburg, MD, USA).2
RSV is divided into types A (RSV-A) and B (RSV-B), based on antigenic differences derived from their glycoproteins G and F, and further categorized into subtypes, based on the variable domain of the attachment G protein. Type A often tends to be more dominant and associated with enhanced disease severity.3 Reinfection by RSV is a common occurrence, because many genotypes can circulate concomitantly. This variability might contribute to the ability of the virus to cause yearly outbreaks and severe disease.4,5 Furthermore, previous reports of RSV infection outbreaks have described varying degrees of disease severity, but have not controlled for the presence of different viral genotypes that may have different virulence.
In Brazil, there is a paucity of data in neonates, regarding the molecular epidemiology of RSV and the emergence of novel viral strains. Although the circulation of NA1 and ON1 RSV genotypes have already been described, the impact on the course of RSV infection in NICUs and possible outbreaks remain to be determined.6–10 The authors report two consecutive RSV outbreaks caused by different genotypes (ON1 and NA2) in a Brazilian NICU. Since early detection of cases is crucial for containment of viral transmission, the study aimed to clarify the importance of molecular detection and typing, as well as infection control measures, in this challenging setting.
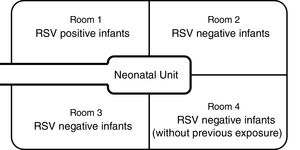
Materials and methodsDesign of the nurserySanta Casa de São Paulo is a tertiary general hospital in downtown São Paulo, with approximately 2000 births annually. The Department of Obstetrics is a referral delivery service for municipal high-risk pregnancy programs, and approximately 15% of newborns require intensive care. The NICU has 28 beds (14 in the intermediate care unit and 14 in the intensive care unit), which are divided among four physically separated wards (Fig. 1). Infants in the main NICU room are kept in either closed incubators or open radiant warmers, and many of them require mechanical ventilation (MV). For those in incubators, most of the routine care is provided through side doors, which are kept closed when not in use. Those in the intermediate care unit stay in open bassinets or closed incubators and do not require MV.
In average, more than 65 different healthcare professionals have physical contact with the babies in the NICU daily: six neonatal hospitalists, eight neonatal fellows, four pediatric residents, four medical school graduation students, six nurse practitioners, 34 nurse technicians, and five respiratory therapists. After hand cleaning and replacement of personal clothing by aprons (provided by the hospital), parents are allowed to visit all wards and to participate in the care of their babies. Children under 18 years of age are not allowed to enter the NICU.
Clinical diagnosisUpper respiratory tract infection was defined when cough and rhinorrhea were present. Bronchiolitis diagnosis was made based on the following symptoms: tachypnea, wheezing, cough, crepitus, and/or nasal flaring.
Outbreak investigationAfter the identification of a symptomatic infant with confirmed RSV infection, nasopharyngeal aspirate (NPA) samples were collected from all infants hospitalized in the NICU. NPA surveillance was repeated twice a week in all patients. The samples were sent to the Institute of Biomedical Science of the University of São Paulo.
Viral RNA was extracted with the automatic NucliSens easyMAG extractor (Biomerieux Inc., Durham, USA), according to the manufacturer's instructions. Real-time reverse transcription (RRT)-polymerase chain reaction (PCR) was performed with the AgPath-ID one-step RRT-PCR kit and the 7300 Real-Time PCR System (Applied Biosystems, Foster City, USA). To identify the pathogen responsible for the outbreak, the RRT-PCR was performed with a non-influenza virus RRT-PCR Panel Kit to detect 15 respiratory viruses – RSV group A and group B, human metapneumovirus, adenovirus, human coronaviruses (HCoV-HKU, -NL63, -OC43, and 229E), human parainfluenza virus (types 1, 2, 3, and 4), human bocavirus, human enterovirus, and human rhinovirus, which were provided by the Centers for Disease Control and Prevention (CDC – Atlanta, GA, USA),11 and commercial primers and probes for influenza A and B (Invitrogen, Carlsbad, USA).
Positive qPCR samples were amplified by conventional two-step PCR for DNA sequencing: cDNA was synthesized using the SuperScript III kit (Applied Biosystems, Foster City, USA) according to the manufacturer's instructions. Analyses of the second hypervariable region of the G protein gene was carried out using PCR with the primers Gr5_fw12 and FVAB_rev13 in a mixture containing 5μL of cDNA, 5μL of 10× PCR buffer, 25mM of each dNTP, 10pmol of each primer, 1.0U of Platinum Taq DNA Polymerase (Invitrogen, Carlsbad, USA), and nuclease-free water for a final volume of 25μL. The second step of semi-nested PCR was performed for negative samples from the first PCR, using the forward primer GR5_fwd and reverse primer F1AB_rev14 in the same conditions as used for conventional PCR. The amplification was performed using the GeneAmp PCR System 9700 thermocycler (Applied Biosystems, Foster City, USA) using the following program: 95°C for 5min, followed by 35 cycles, each consisting of 30s at 95°C, 30s at 55°C, and 45s at 72°C, and finally 5min of extension at 72°C. The amplified products were analyzed using agarose gel electrophoresis and visualized under ultraviolet light after staining with ethidium bromide. The amplified products of gene G of ∼1052 and 900bp, produced using conventional PCR and semi-nested PCR, consecutively, were purified using ExoSap-IT (Affymetrix Inc., Santa Clara, USA) and submitted to a cycle sequencing reaction (Sanger) using a fluorescent dye terminator kit with all primers, GR5, FV, and F1AB, in the 3100 DNA Sequencer (Applied Biosystems, Foster City, USA). Both strands of each amplification product were sequenced at least twice. Sequence editing, alignments, and phylogenetic analyses were performed with the MegAlign 5.03v software (DNAStar Inc., Madison, USA). Standard published sequences from subgroup A were downloaded from GenBank (https://www.ncbi.nlm.nih.gov/genbank) as a reference of different lineages and genotypes.
EthicsThe study was approved by the local research ethics committee.
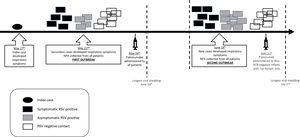
ResultsFirst outbreak (May)The index case involved a 30-day-old preterm boy (33 weeks of gestational age) with a birth weight of 2290g who had been hospitalized since birth because of prematurity and bronchopulmonary dysplasia. He was being weaned off oxygen supplementation in the intermediate care unit when cough, runny nose, and fever started. Respiratory failure developed rapidly, requiring MV. Five days later, five infants (out of 17 who were in the NICU) developed respiratory symptoms; therefore, an outbreak was declared (Fig. 2). On the same day, NPA samples were collected from all patients in the NICU for RRT-PCR respiratory virus detection. Ten infants were infected with RSV, of whom only three shared the ward with the index case. The attack rate was 58.8%, and only one fatality occurred (CFR=10%). However, the death was not attributed to RSV infection, since the patient did not present with respiratory symptoms and had congenital pulmonary hypoplasia, which was considered the cause of death. Additional information is provided in Table 1.
Clinical characteristics of patients infected by RSV in the first outbreak (caused by ON-1 genotype) and second outbreak (caused by NA-2 genotype).
| 1st outbreak: RSV-A type ON-1 (n=10) | 2nd outbreak: RSV-A type NA-2 (n=14) | |
|---|---|---|
| Characteristics | ||
| Gestational age (mean, weeks) | 34.9 | 33.4 |
| Birth weight (mean, g) | 2.327 | 1.700 |
| Age at onset (mean, days) | 35.4 | 14.3 |
| Comorbidities | ||
| Congenital heart disease | 2 (20%) | 1 (7.1%) |
| Lung disease | 1 (10%) | – |
| Prematurity (<37 weeks) | 5 (50%) | 11 (78.6%) |
| Concurrent or previous late-onset sepsis | 3 (30%) | 5 (35.7%) |
| Clinical presentation and outcomes | ||
| Asymptomatic | 3 (30%) | 8 (57.1%) |
| Mild upper respiratory infection | 2 (20%) | 1 (7.1%) |
| Bronchiolitis without MV | 4 (40%) | 1 (7.1%) |
| Bronchiolitis with MV | 1 (10%) | 4 (28.6%) |
| Deaths | 1 (10%) | 2 (14.3%) |
RSV, respiratory syncytial virus; Upper respiratory tract infection: presence of cough, rhinorrhea; Bronchiolitis: tachypnea, wheezing, cough, crepitus and/or nasal flaring; MV, mechanical ventilation.
Contact and respiratory precautions were immediately implemented. All infants in the NICU were separated into three different cohorts. RSV-positive infants were placed in ward 1, under contact and respiratory precautions. RSV-negative infants who had previous contact with any of the RSV-positive infants were placed in wards 2 and 3, under contact precautions only. RSV-negative infants who had never had contact with RSV-positive patients were placed in ward 4.
Healthcare staff was also divided across the wards, and their internal rotating schedules were suspended. Fifty of them had NPA and saliva specimens tested for respiratory virus, which were negative. NICU visitation was restricted and its duration was reduced. Infection control measures were reinforced for healthcare staff and infants’ families by the infection control group and NICU members, and they included hand hygiene, environmental cleaning, and increased awareness of respiratory symptoms. On the second day of the outbreak, palivizumab was administered to all infants in the NICU.
NPA surveillance (testing for the 17 respiratory viruses as previously described) was performed twice a week in all infants in the NICU so that viral shedding duration and possible co-infections could be determined. Follow-up continued for four weeks after the last positive sample was identified. RSV-positive infants were kept isolated until NICU discharge and were tested weekly using PCR until a negative result was achieved. The longest viral shedding period was 29 days.
Second outbreak (June)During the following 24 days, no other infants in the NICU developed symptoms suggestive of RSV infection; therefore, the first outbreak was considered controlled. Thirty-four days after the index case, a new outbreak was identified, and control measures were reinstated (Fig. 2). Fourteen infants had RSV identified in respiratory secretions (Table 1). Palivizumab was administered only to RSV-PCR-negative infants with risk factors (eight patients). In this second outbreak, the attack rate was 63.6%, and two deaths occurred (CFR=14.3%). Both infants were premature and were receiving MV when RSV infection was detected. Their deaths were attributed to bacterial sepsis; however, RSV could have contributed. The longest viral shedding time was 14 days.
Characteristics of infected infantsAmong the 24 RSV-positive infants, 15 (62.5%) were born prematurely at less than 37 weeks of gestation (Table 1). The mean gestational age was 33 weeks (range: 28–40 weeks), mean age at diagnosis was 27.5 days (range: 1–80 days), and mean birth weight was 1961g (range: 765–3440g). A total of 11 infants (45.8%) were asymptomatic. Among the symptomatic infants, 12.6% presented with mild upper respiratory symptoms (cough or coryza) or fever, 20.8% presented with bronchiolitis without the need for MV, and an additional 20.8% required invasive ventilatory support. Six infants would be eligible by the Brazilian guidelines to receive palivizumab.15 In both outbreaks, the available data were not sufficient to establish independent risk factors for RSV infection.
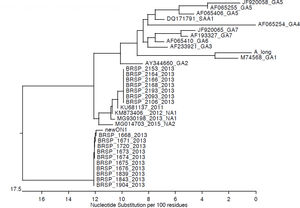
Genotypic characterizationMolecular analysis was performed for all 24 RSV-positive samples (Fig. 3). Viruses isolated during the first outbreak were characterized as RSV-A type ON-1, while all infections during the second outbreak were caused by RSV-A type NA-2. Aminoacid analysis evidenced a characteristic genetic signature in the second outbreak samples. No viral coinfection was detected in either outbreak.
DiscussionTo the best of the authors’ knowledge, this is the first report of two consecutive neonatal outbreaks caused by two distinct genotypes of RSV in Brazil. Together, the episodes affected 24 infants in a high complexity NICU and occurred in the RSV post-peak period (the highest viral circulation rates occur between March and May in São Paulo). The RRT-PCR assay and genetic sequence analysis were pivotal in the confirmation of the two distinct outbreaks and the adoption of control measures, including palivizumab.
Preterm infants with underlying heart or pulmonary disease are more prone to developing severe symptoms when infected with RSV. Compared with community-acquired infections, nosocomial infections are associated with significantly higher mortality.2 The attack rates among RSV outbreaks in NICU reported in the literature vary from 9 to 55%.16–21 The attack rates observed in the present study were much higher, reaching 58.8% during the first outbreak and unexpectedly reaching 63.6% in the second. Such disparity could be attributed to the fact that most of the present patients were presumably naïve to the new genotypes described and that none had received palivizumab before the onset of the outbreak. The absence of protective immunity in the infants and their mothers could allow more efficient virus replication and transmission. Furthermore, such emergent genotypes appear to have a fitness advantage over preceding genotypes of the same RSV group.22
In contrast, the authors did not observe an increased frequency of severe disease because only 10% of the patients in the ON-1 group and 28.6% in the NA-2 group developed respiratory failure and required MV. Such frequency could be explained by the relatively high mean gestational age (34 weeks) of the present patients. Although no extremely preterm infants were included, all five RSV-infected infants who required MV were born prematurely at less than 33 weeks, reinforcing the contribution of prematurity as a major risk factor for severe RSV infection.
There are no guidelines for the use of palivizumab during outbreaks in healthcare settings. Small studies that used palivizumab in NICUs as a component of outbreak control procedures were retrieved in the literature.17–20 When the first outbreak was detected in this setting, all 17 patients in the NICU received palivizumab, while palivizumab was available only to RSV-negative patients in the second outbreak due to the limited amount of medication available and its high cost. Although the minority of infants had a strict indication of receiving palivizumab, the authors believe the broad and prompt administration of the drug may have contributed to the timely control and the low morbidity/mortality in the studied population.
Previous studies described the co-circulation of RSV groups A and B in epidemic periods, including in Brazilian populations.5,7,9,23,24 RSV-A was responsible for both outbreaks; however, our cases occurred in a closed unit. Nevertheless, two different RSV-A genotypes were identified in the NICU: ON-1 and NA-2. The first is a novel genotype with large duplications of amino acids in the attachment G glycoprotein. It was first reported in December 2010 in Ontario, Canada and has been rapidly spreading globally.22,25–30
The occurrence of RSV and specific genotypes during nosocomial outbreaks may mirror the circulation of the virus in the community. This is partially demonstrated by the time of these outbreaks, which respect the seasonality in São Paulo.
In summary, early etiologic diagnosis is especially relevant for antiviral therapy and isolation measures to prevent hospital outbreaks, reducing the need for additional diagnostic procedures and length of hospital stay. A better understanding of RSV molecular epidemiology is essential for vaccine and antiviral drug development, and other control measures against RSV infection.
Conflicts of interestThe authors declare no conflicts of interest.
Please cite this article as: Silva DG, Almeida FJ, Arnoni MV, Sáfadi MA, Mimica MJ, Jarovsky D, et al. First report of two consecutive respiratory syncytial virus outbreaks by the novel genotypes ON-1 and NA-2 in a neonatal intensive care unit. J Pediatr (Rio J). 2020;96:233–9.