To discuss the recent literature on paternal obesity, focusing on the possible mechanisms of transmission of the phenotypes from the father to the children.
SourcesA non-systematic review in the PubMed database found few publications in which paternal obesity was implicated in the adverse transmission of characteristics to offspring. Specific articles on epigenetics were also evaluated. As the subject is recent and still controversial, all articles were considered regardless of year of publication.
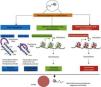
Summary of findingsStudies in humans and animals have established that paternal obesity impairs their hormones, metabolism, and sperm function, which can be transmitted to their offspring. In humans, paternal obesity results in insulin resistance/type 2 diabetes and increased levels of cortisol in umbilical cord blood, which increases the risk factors for cardiovascular disease. Notably, there is an association between body fat in parents and the prevalence of obesity in their daughters. In animals, paternal obesity led to offspring alterations on glucose-insulin homeostasis, hepatic lipogenesis, hypothalamus/feeding behavior, kidney of the offspring; it also impairs the reproductive potential of male offspring with sperm oxidative stress and mitochondrial dysfunction. An explanation for these observations (human and animal) is epigenetics, considered the primary tool for the transmission of phenotypes from the father to offspring, such as DNA methylation, histone modifications, and non-coding RNA.
ConclusionsPaternal obesity can induce programmed phenotypes in offspring through epigenetics. Therefore, it can be considered a public health problem, affecting the children's future life.
Discutir a literatura recente sobre obesidade paterna, focalizando os possíveis mecanismos de transmissão dos fenótipos do pai para os filhos.
FontesUma revisão não-sistemática no banco de dados PubMed encontrou poucas publicações com obesidade paterna implicada com a transmissão adversa das características à prole. Artigos específicos sobre epigenética também foram avaliados. Como o assunto é recente e ainda controverso, todos os trabalhos foram considerados independentemente do ano de publicação.
Resumo dos achadosEstudos em seres humanos e animais estabeleceram que a obesidade do pai prejudica seus hormônios, metabolismo e função espermática, que pode ser transmitida à prole. Em humanos, a obesidade paterna resulta em resistência à insulina/diabetes tipo 2 e aumento do nível de cortisol no sangue do cordão umbilical, que aumenta os fatores de risco para doença cardiovascular. Notavelmente, existe associação entre a gordura corporal nos pais e a prevalência de obesidade em suas filhas. Em animais, pais obesos condicionam, na prole, a homeostase glicose-insulina, lipogênese hepática, hipotálamo/comportamento alimentar, rim, prejudicam o potencial reprodutivo da prole masculina com estresse oxidativo espermático e disfunção mitocondrial. Uma explicação para estas observações (humanos e animais) é a epigenética, considerada a ferramenta básica para a transmissão de fenótipos do pai à prole, como a metilação do DNA, modificações nas histonas, e RNA não codificante.
ConclusõesA obesidade paterna pode induzir fenótipos programados na prole através da epigenética. Portanto, a obesidade paterna pode ser considerada um problema de saúde pública, afetando a vida futura das crianças.
Obesity has been growing in a disorderly way, constituting a real epidemic described as “globesity,” which represents a serious public health problem nowadays.1
It is now known that the risk of developing obesity and metabolic syndrome (MS) in adulthood may be influenced by the initial period of life, especially through inadequate nutrition available to the fetus and newborn.2,3 “Programming” is the process by which early life factors may influence the offspring's health in adulthood. Programming is considered an essential mechanism for the establishment of obesity and metabolic changes in the offspring.4,5 Various models are used to understand the mechanisms associated with programming, in which the hormonal and metabolic environment during the prenatal or postnatal period is altered through changes in maternal nutritional status.6–8
Most epidemiological and experimental studies have focused on the maternal influence on offspring's health. However, recent experiments with rodents have demonstrated that the paternal involvement affects glucose homeostasis and the lifetime of pancreatic islets in female offspring.9 Clinical and animal testing have challenged conventional ideas about metabolic programming, suggesting that something else might act in this process via paternal programming. Recent studies now indicate that paternal metabolic health at conception can also impact children's health, and that obese fathers are more likely to generate an obese child.10
This review reports the recent findings and proposed mechanisms involved in paternal programming of the offspring.
Human studiesStudies in humans analyzed the relationship between paternal lifestyle-related factors, environmental exposure factors, and offspring's health outcome in early and later life, suggesting that paternal effects may play a significant role in the pathogenesis of offspring chronic diseases in later life (e.g., insulin resistance and type 2 diabetes). More than 60% of all adults are classified as overweight or obese in most Westernized societies; as the prevalence of obesity increases, it is responsible for an ever-larger proportion of the overall burden of disease.9,11
There is clear evidence that paternal nutritional factors play a significant role in the health of offspring. For example, there is a correlation between paternal absolute and relative amounts of body fat and the same parameters in their daughters aged 4.8–8.9 years.12 Furthermore, paternal body mass index (BMI) might modulate the offspring phenotype in a sex-dependent manner. In the frames of a family cohort study (899 parent–offspring trios), paternal BMI was correlated with birth weight, biparietal diameter, head circumference, abdominal diameter, abdominal circumference, and thoracic diameter in male newborns only.12 Fathers13 or grandfathers14 exposed to either overfeeding or food restriction on the age period of 9–12 years predetermined reduced longevity of their male offspring. The second generation of offspring of these grandfathers had a four-fold risk of diabetes mortality.15
In Northern Sweden, the follow-up of three generations demonstrated that a grandfathers’ surfeit of food is associated with reduced survivability14 and an increased risk of diabetes15 in their grandchildren. Early onset of grandpaternal smoking is also related to increased grandson BMI.16 This evidence indicates that paternal nutritional factors, not only before conception but also as early as in father's puberty, might affect the offspring in a sex-dependent manner.17 Moreover, there is an interaction between parental genes and parental environmental factors that affect the phenotype of the offspring.18 The gene–environment interaction becomes even more complicated, as it is also known that the socioeconomic status of an individual appears to have opposite effects on obesity in poor and rich countries.19
In fathers, the caloric imbalance imposed by lifestyle choices, including high food consumption and low physical activity, are factors to be considered in programming studies. Epigenetic modifications can occur within the lifespan of numerous individuals within a population, and thus be transmitted immediately to a large number of offspring in the next generation, unlike genomic events that spread slowly through a population.20 It is likely that changing circumstances within the individual or over several generations can recruit silent alleles back into the active genome and contribute to the reversibility of adaptive or acquired changes. A recent study in obese men showed changes in circulating microRNA (miRNA) that target VEGF (vascular endothelial growth factor), an adipocyte mitogen, which was reversible following weight loss.21
Paternal BMI during conception was associated with fetal development of the male offspring, but not of the female offspring. It was significantly correlated with birth weight and perinatal biparietal diameter, head circumference, abdominal diameter, abdominal circumference, and thoracic diameter measured in male offspring. There were no significant correlations between paternal BMI and these parameters in female offspring. Cord blood cortisol level was also associated with father's BMI in male offspring only. The authors concluded that a sex-specific transgenerational effect of paternal BMI on fetal cortisol secretion might represent a risk factor for cardiovascular disease in male offspring in later life.12 Furthermore, increased paternal BMI is associated with decreased blastocyst development and live birth rates after in vitro fertilization.22 In obese fathers (BMI>25kg/m2), a higher reactive oxygen species was detected in sperm, as well as increased seminal fluid neopterin (a marker of reproductive tract macrophage activation), decreased sperm counts and serum testosterone, and increased serum estradiol.23
Animal studiesAnimal models of male obesity are being used to assess the impact of paternal programming on offspring and to analyze the sperm function of the obese father. Animal models are important due to the difficulties in separating the effects of paternal genetic makeup from those of paternal environmental exposures on the offspring,10 as well as in clustering and interpreting data from human studies. A better understanding of the mechanisms of paternal programming can help in interventions to minimize adverse effects on the offspring.24
A paternal programming (when obese fathers led to disturbance of glucose-insulin homeostasis in the female offspring) was initially described in animals.9 Obese fathers also program liver lipogenesis and beta-oxidation25 and the hypothalamus of the offspring (hypothalamus inflammation was found in offspring, with an increase of interleukin [IL]-6 and tumor necrosis factor [TNF]-alpha expressions).6
Obese fathers also altered the offspring's kidney, with tubular damage and loss of the tubular brush border, but not glomerular damage. The cholesterol acyltransferase-1 (Acat1) gene, involved in the input of fatty acid for beta-oxidation in the tubuli, was up-regulated in the offspring.26
Mammalian male germ cell development is susceptible to damage in different times and in specific diseases in offspring's later life: embryonic development, infancy, and prepubertal age, and preconception and spermatogenesis in adulthood.27–29 Paternal obesity negatively impacts on the reproductive potential of the male offspring not only by altering function, quality, and molecular composition of sperm, but also by increasing sperm's oxidative stress, contributing to DNA damage and mitochondrial dysfunction.30
Many studies in animal model focus on the adverse factors influencing paternal exposure, from the prepubertal period to preconception. Therefore, is more plausible to consider that the epigenome undergoes reprogramming in pre-implantation embryos and in primordial germ cells.31
Environmental exposures in animal models over this period have been suggested to induce intergenerational and transgenerational effects through the sperm epigenome.32 Data demonstrate that numerous metabolic effects observed in the first generation are shown to persist in the second generation, with F1 females producing F2 males with increased adiposity. Paternal obesity modified the expression of various microRNAs, concomitant with alterations in sperm microRNA content and a reduction in global methylation of germ cell DNA.32
From the moment the blastocyst is formed, the pre-implantation is over, and a new stage starts – the in utero development. After entering the gonads, primordial germ cells convert to gonocytes, and in mouse models decreasing decrease in DNA methylation levels is observed.33 The primary responsibilities in this timeframe encompass cell-specific gene expression, tissue differentiation, and tissue-specific epigenomes.27
Obese mice fathers presented H3 retention and genomic imprints in the sperm, and differences in liver mRNA expression of several fat synthesis-related genes were observed in the offspring. Differences were observed in the liver expression of matallothionein-1 and -2 (Mt1 and Mt2), fatty acid synthase (Fasn), P450 cytochrome oxidoreductase,34 and acetyl-CoA carboxylase-α (Acaca) at the age of 24 weeks. Paternal obesity also increased histone H3 occupancy in the promoters of the genes responsible for the embryonic development and increased monomethylation of lysine 4 on histone H3 (H3K4me1) in genes responsible for embryogenesis regulation in spermatozoa. Altogether, findings suggest that dietary exposure can modulate histone composition at genes involved in the process of development.35
EpigeneticsThe mechanisms explaining how the father may affect the development of the offspring are still under debate. Epigenetics is the primary tool for the transmission of paternal phenotypes to offspring, because the transient nutritional stimuli in critical stages of ontogenesis may influence the expression of various genes by changes in chromatin conformation and the accessibility of transcription factors.36
Therefore, the term epigenetic (Greek prefix epi- (¿π¿): over, outside of, around), proposed by Conrad Waddington, is described as a “process of development of the phenotype from the genotype.”37 In other words, epigenetic is any transmissible and reversible modification in the expression of a gene without structural change in the sequence of DNA.38 Unlike the genetic variation of the germline, which remains unchanged in all cells of the body, epigenetic modification is dynamic and varies among tissues in response to a range of environmental stimuli, including those that direct tissue differentiation during development and growth, and the serious risks that provoke an adaptive response of cells.39
Some epigenetic marks during spermatogenesis may continue throughout the embryonic development. Environmental exposures (diet, lifestyle, and other exposures) that occur in male gametogenesis can cause irreversible epigenetic changes and phenotypic consequences expressed in the offspring.40 Epigenetic processes also modulate the effects through transcription regulation due to several processes, such as DNA methylation, histone alterations, and transcription of non-coding RNA (miRNA, for example).41,42
Epigenetic modifications acting on the cell plasticity capacity prepare the individual for the extrauterine environment and may potentiate a survival advantage by regulating the differential genes encoding proteins involved in energy metabolism and adipogenesis.43 However, in face of a deleterious metabolic condition such as obesity and related metabolic alterations, these modifications can be exacerbated or silenced, especially germ cells program, which constitute the species’ perpetuation and phenotypic transmission.44
Recent hypotheses consider that some paternal dietary patterns reside in spermatozoa bearing epigenetic information.27 The sperm “epigenome” was traditionally considered insignificant, since it was postulated that its DNA methylation profile was erased immediately after fertilization. However, in recent years, there has been an increase in the number of reported cases of apparent epigenetic inheritance through the male germline, suggesting that this epigenome may transmit information between generations.28,40
The development of sperm- and spermatid-derived frog embryos allow the observation that the sperm not only transfers DNA, but also contributes to the epigenetic information required for proper embryonic gene expression, being the key for epigenetic information.45
MethylationDNA methylation is one of the chemical reactions that occur most frequently in eukaryotes such as plants, fungi, invertebrates, and vertebrates.34 This chemical modification is characterized by the addition of a methyl group at the C5 position of the cytosine ring, catalyzed by the DNA methyltransferases, leading to the formation of 5-methylcytosine.46,47 The frequency of 5-methylcytidine is less than 1% of the total number of nucleotides in the genome.48 This methylation process acts in normal embryonic development, inactivation of the X chromosome, genetic regulation, genomic press, and chromatin modifications.49
Most DNA methylation occurs in regions called CpG islands, which corresponds to genomic regions of more than 1000 base pairs in length and with many GC dinucleotides; about 55% of these islands are in the promoter regions of approximately 40% of mammalian genes. These CpG (cytosine-phosphate-guanine) islands are kept non-methylated, except in imprinting genes or when located on the inactive X chromosome.50,51 Therefore, CpG islands located in the promoter region of housekeeping and developmental regulator genes with dense cytosine and guanine distribution are resistant to DNA methylation.52 The CpG islands, which allow the binding of proteins and enzymes, initiate the cascade of transcription. In contrast, methylated CpG islands are related to transcriptional silencing.53
Hypermethylation of promoter regions rich in dinucleotides has a significant role in the loss of gene expression.54 Typically, hypomethylation of DNA triggers an increase in gene expression, while hypermethylation decreases the expression of target genes.55 Furthermore, transcription factors do not recognize and bind to transcription initiation sites due to the modification of cytosine into 5-methylcytidine. This is the case of adipocyte protein 2 (AP-2), cMYC/murine homolog of max (cMYC/MYN), cyclic adenosine monophosphate response element binding protein (CREB), E2 factor (E2F), and nuclear factor-κB (NF-KB). However, these binding sites may be occupied by other proteins such as methyl-CpG binding protein 2 (MeCP-2) and methyl-CpG-binding domain protein (MBD)1, MBD2, MBD3, and MBD4, which bind to methylated cytosines and stimulate chromatin condensation, inactivating the gene.46,56
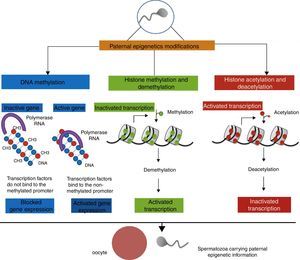
The enzymes responsible for the addition of a methyl group to cytosine molecules belong to the family of DNA methyltransferases (DNMTs), including DNMT-1, DNMT3A, DNMT3B and its isoforms, and DNMT3L.47,57 DNMT1 is primarily responsible for maintaining DNA methylation patterns during mitosis. Furthermore, DNMT (differentially methylated regions)-s3 is responsible for de novo methylation of newly synthesized DNA molecules and is most important during the early stages of embryonic development; it could be the process involved in paternal programming for phenotypic transmission.58,59 A schematic view of the main paternal epigenetic modifications can be seen in Fig. 1.
Epigenetics of paternal programming on the offspring. Paternal metabolic changes related to obesity may result in modifications to genetic information contained in the spermatozoa. DNA methylation in specific promoter regions most often prevents gene transcription by inactivating the gene in question. As for histone modifications, hypermethylation (depending on histone and amino acid) favors the condensation of chromatin, making difficult access to regulatory proteins that promote transcription. However, non-methylated histones ensure decondensed chromatin, supporting gene transcription. In contrast, histone acetylation opens the chromatin, allowing coupling of the transcriptional machinery.
In a contrary process, DNA demethylation is also an important epigenetic component of gene transcription and epigenetic programming, which occurs through several enzymatic reactions mediating the oxidation of 5-methylcytosine to 5-hydroxymethylcytosine.60 The presence of 5-hydroxymethylcytosine in promoter regions results in increased transcription, indicating a role in long- and short-term regulation of gene expression.61
Histone modificationsHistone modifications are other biological actions that regulate gene expression. Histones are basic proteins found in eukaryotic cell nuclei, which help in the packaging of DNA into nucleosomes, the binding blocks of chromatin.62 Chromatin should be remodeled, modifying the accessibility of the DNA transcription machinery to control the transcription process.63 Events such as acetylation, methylation, phosphorylation and ubiquitination, often occur in the tail of the histones that extend from the center of the nucleosomes.64 Other chemical modifications also alter histones, such as histone lysine glucosamine acylation (GlcNAcylation), butyrylation, malonylation, and crotonylation.65,66
Histones 3 and 4 (H3 and H4) are commonly studied, and acetylation is the main epigenetic modification considered; it occurs on the lysine and arginine and neutralizes the positive charge of basic residues. The histone acetylase enzymes add acetyl groups to the histone lysine residues, and it is believed that the acetylated histones have a reduced affinity between DNA and histones, leaving chromatin in a relaxed (euchromatin) and transcriptionally active state.66
In contrast, histone deacetylase removes the acetyl groups, which are more condensed, and prevents gene expression.67 These chemical modifications alter the interaction between DNA and histones, changing the degree of chromatin folding and the gene activity.68 Therefore, heterochromatin is related with hypoacetylation for H3 and H4, and di- or trimethylation of the ninth lysine residue on H3 (H3K9me2 or H3K9me3).69
Non-coding RNA (microRNA)In addition to DNA methylation and histone modification, sperm RNA can be an epigenetic regulator. Spermatozoa contain an array of both mRNAs70 and noncoding RNAs, including miRNA.71 Most of this RNA is delivered to the oocyte. However, the role of miRNA in early preimplantation embryos is still under debate.
Evidence of the direct biological effect of miRNAs in the pre-implantation period is supported by observations in mice with a chromosomal lesion in the Dicer gene. Loss of enzymatic processing of Dicer miRNA in oocytes leads to early lethality in the development process, where the zygotes cannot survive the division in the first place.72 Therefore, this suggests the interference of miRNA in zygote development.
miRNA is a major part of the group of such small non-coding. These components have approximately 21nt-long RNA molecules that repress their target mRNA.73 In males, they are observed in the nucleus of spermatozoa, keeping the DNA connected to histones during spermiogenesis and in early embryonic development.74 miRNA regulates various biological functions, influencing the epigenetic inactivation of genes and the protection of the DNA against viruses and transposons.75
In a general way, miRNA in animals is mostly located within the introns of protein coding or non-coding RNA genes,76 being produced by RNA polymerase II transcription. In humans, miRNA appears to be synthesized by RNA polymerase III.77 In recent years, over- or under-expression of miRNA has been associated with disease development, but the mechanisms and actions are still ambiguous.78
Epigenetics in paternal programmingSeveral studies in animals and humans have demonstrated the influence of paternal diet on their offspring's phenotype through the epigenome.
Male mice fed a high-fat diet generated female offspring with impaired glucose-insulin homeostasis, associated with an altered expression in one of the 642 pancreatic islet genes and in the hypomethylated interleukin 13 receptor alpha 2 (IL13ra2) gene.9 These alterations were recently observed in the transcriptomes of retroperitoneal adipose and pancreatic islet tissues in female offspring. In retroperitoneal adipose tissue, 5108 genes were differentially expressed due to paternal high-fat diet, whose functions are related to mitochondrial and cellular response to stress, telomerase signaling, cell death and survival, cell cycle, cellular growth and proliferation, and cancer.79
In a mice model of paternal low-protein diet, the offspring showed an elevated methylation in peroxisome proliferator-activated receptor (PPAR)-alpha in the liver, a gene involved in the formation of the first enzymes for the oxidation of lipids in mitochondria, being an essential lipid regulator.80 Also, paternal insulin resistance has altered the methylation status of various insulin-signaling genes in the offspring, increasing the offspring's susceptibility to diabetes through gametic epigenetic alterations. Such includes the genes phosphoinositide-3-kinase regulatory subunit 1 (Pik3r1), phosphoinositide-3-kinase catalytic alpha polypeptide (Pik3ca), Ptpn1 protein tyrosine phosphatase, non-receptor type 1 (Ptpn1), and Pik3ca in sperm.81 In mice models, paternal high-fat diet led to an increase in transfer RNA (tRNA) in sperm as an inherited epigenetic key influenced by paternal diet and related to metabolic impairment in the offspring, leading to glucose intolerance and insulin resistance.82
Paternal obesity initiates metabolic disturbances in two generations of mice, altering the transcriptional profile of testis and sperm miRNA content. The differential content of canonical miRNAs in the sperm suggested dysregulation in spermatogenesis, embryo development, and metabolic function. Furthermore, paternal high-fat diet affected the metabolic status of offspring through epigenetic changes in the adiponectin and leptin genes for two generations.83 Interestingly, an increase in miR-29 (microRNA 29) has been associated with a decrease in methylation of repeat elements in the male germline.32 Moreover, 13 sperm-borne miRNA were modulated by paternal high-fat diet and transferred off this altered miRNA payload to the embryo at fertilization, changing its growth trajectory and affecting adult offspring phenotype.84
In newborns, an association was observed between preconception obesity and DNA methylation profiles in the offspring, particularly in the differentially methylated region (DMR) of the imprinted insulin-like growth factor 2 (IGF2) gene. The hypomethylation at the IGF2 DMR in the offspring was associated with paternal obesity.85 It is conceivable that additional or novel epigenetic regulators (such as prions) are present in sperm, that sperm quality is affected by diet, and that environment may direct the genetic changes. Although it is important to emphasize that inbred mouse strains were used in this study.80 Furthermore, γ-radiation-induced DNA damage in the sperm has been shown to be heritable, with offspring similarly exhibiting sperm DNA damage.86
Epigenetic modification is a continual process, and some changes may be reversible.20 Combined, these data reveal the impact of paternal programming (in particular through nutritional manipulations) on the future life of the offspring, influenced mainly by phenotype transmission via the epigenetic process.
Final considerationsData from epidemiological and animal studies provide evidence that paternal feeding and paternal health conditions can program following generations. Thus, the mother is not the only responsible for offspring's health. The father shares the responsibility in providing sperm-specific epigenetic stamp to the oocyte, affecting the embryo developmental trajectory and health of adult offspring. Although the role of paternal influence may be clearly identified, the current knowledge on reprogramming, alteration, and possible prevention of paternal effects remains limited. To date, human studies are not progressing in the same manner as animal studies, so there is little information about the mechanism and contribution of paternal programming in the child's health, as men are widely used as controls in studies with women. Therefore, considering that paternal obesity can also be a public health problem, it is important to improve epidemiological studies to assess the exact role of paternal health on the sperm and on the health of the offspring to propose further interventions.
Conflicts of interestThe authors declare no conflicts of interest.
This laboratory (www.lmmc.uerj.br) is sponsored by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, grant #302.154/2011-6 to CAML, and #306.077/2013-2 to MBA), and Fundação Carlos Chagas Filho de Amparo à Pesquisa do Rio de Janeiro (FAPERJ, grant #102.944/2011 to CAML, #103.062/2011 to MBA).
Please cite this article as: Ornellas F, Carapeto PV, Mandarim-de-Lacerda CA, Aguila MB. Obese fathers lead to an altered metabolism and obesity in their children in adulthood: review of experimental and human studies. J Pediatr (Rio J). 2017;93:551–9.