Since the present group had already described the composition of the intestinal microbiota of Brazilian infants under low social economic level, the aim of the present study was to analyze the microbial community structure changes in this group of infants during their early life due to external factors.
MethodsFecal samples were collected from 11 infants monthly during the first year of life. The infants were followed regarding clinical and diet information and characterized according to breastfeeding practices. DNA was extracted from fecal samples of each child and subjected to Polymerase Chain Reaction - Denaturing Gradient Gel Electrophoresis.
ResultsThe results revealed a pattern of similarity between the time points for those who were on exclusive breastfeeding or predominant breastfeeding. Although there were changes in intensity and fluctuation of some bands, the Denaturing Gradient Gel Electrophoresis patterns in the one-year microbial analysis were stable for breastfeeding children. There was uninterrupted ecological succession despite the influence of external factors, such as complementary feeding and antibiotic administration, suggesting microbiota resilience. This was not observed for those children who had mixed feeding and introduction of solid food before the 5th month of life.
ConclusionThese results suggested an intestinal microbiota pattern resilient to external forces, due to the probiotic and prebiotic effects of exclusive breastfeeding, reinforcing the importance of exclusive breastfeeding until the 6th month of life.
Como nosso grupo já havia descrito a composição da microbiota intestinal de neonatos brasileiros em baixo nível socioeconômico, o objetivo deste estudo foi analisar alterações estruturais da comunidade microbiana deste grupo de neonatos no início de sua vida devido a fatores externos.
MétodosAmostras fecais foram coletadas mensalmente de 11 neonatos durante o primeiro ano de vida. Os neonatos foram acompanhados com relação a informações clínicas e nutricionais e caracterizados de acordo com práticas de amamentação. O DNA foi extraído das amostras fecais de cada criança e submetido a análise através da técnica de Reação em Cadeia da Polimerase - Eletroforese em Gel de Gradiente Desnaturante.
ResultadosOs resultados revelaram um padrão de similaridade entre seus próprios pontos temporais em indivíduos em aleitamento materno exclusivo ou predominante. Apesar de variações na intensidade e flutuação de algumas bandas, o padrão Eletroforese em Gel de Gradiente Desnaturante na análise microbiana de um ano foi estável em crianças em aleitamento materno. Houve sucessão ecológica ininterrupta apesar da influência de fatores externos, como alimentação complementar e administração de antibióticos, sugerindo resiliência da microbiota. Isso não foi observado nas crianças com alimentação heterogênea e introdução de alimentos sólidos antes do quinto mês de vida.
ConclusãoNossos resultados sugerem um padrão de microbiota intestinal resiliente a forças externas, devido a efeitos probióticos e prebióticos do aleitamento materno exclusivo, reforçando a importância do aleitamento materno exclusivo até o sexto mês de vida.
The intestinal microbiota is an important key in health and disease.1 It is well known that the intestinal microbiota compositions of newborns and children are influenced by birth,2 diet,3 geographic region, and environmental influences.4,5
New methods in 16S rRNA sequencing6,7 are rewriting the understanding about the relationship between bacteria and human host, and the use of these methods for characterizing the intestinal microbiota of infants and children living in developed countries has received increasing attention.3,8 However, few studies conducted in developing countries have corroborated the global observation of inter-individual variability,4,9 despite the differences in intestinal microbiota composition. All these findings may contribute to the worldwide understanding of how a geographic region and its environmental contamination influence or do not influence the intestinal microbiota establishment.
The authors previously described the microbiota composition of a group of children living at a low socioeconomic level in Brazil, characterized by low rates of Staphylococcus in early ages10 and a high abundance of Escherichia at the 12th month of age.5 However, information about how the microbial community structure changes in the short term due to external factors was missing.
Denaturing gradient gel electrophoresis (DGGE)11 has demonstrated that the dynamics of the diversity in the intestinal microbial community can be assessed to determine microbial structural differences between environments12 and their changes over time.13 The present study analyzed the microbial community structure of this group of children monthly during their first year of life, using DGGE.
Material and methodsSubjects and samplesA group of 11 children living at a low socioeconomic level was analyzed, as previously described.5,10 The infants were vaginally delivered at the University Hospital of the University of São Paulo (HU-USP). Information concerning the socioeconomic condition of the family and sanitary conditions was collected monthly, during medical appointments. The child's diet was monitored regularly, concerning the period of lactation and timing of the introduction of new foods. The occurrence of infections, health complications, and medication uses were recorded. Fecal samples were collected from babies on the first 2 days, the 7th day, and every month up to the 12th month of life. The mothers were instructed to collect the fecal sample immediately after elimination with a standardized sterile spoon, place it in a sterile plastic container, and keep it in a freezer (−20°C) until the appointment some hours later. Samples were transported all the way to the laboratory in an ice-filled polystyrene container, numbered (Table 1S – supplementary material) and immediately stored at −80°C until DNA extraction, performed in two to three weeks after the collection.
Breastfeeding practicesThe children enrolled in this study were characterized according to feeding practices, based on WHO Indicators for Assessing Breastfeeding practices,14 as follows: exclusive breastfeeding for those children who received only breast milk; predominant breastfeeding for those who received breast milk, water and water based drinks, and fruit juice; and complementary feeding for those who received breast milk and any food including non-human milk (Table 1). Also, according to the WHO indicators,14 the time recommended for exclusive breastfeeding was up to the 6th month of age.
Clinical information on antibiotic use and breastfeeding practices.
| Child # | Gender | Antibiotic use (time) | Breastfeeding practicea |
|---|---|---|---|
| 1 | F | Amoxicillin (9th month – 7 days) | Complementary feeding |
| 2 | F | NO | Complementary feeding |
| 3 | F | Cephalexin (1st month – 7 days) | Predominant breastfeeding |
| 6 | M | Amoxicillin(7th m month – 10 days) | Complementary feeding |
| 7 | F | NO | Complementary feeding |
| 8 | F | Amoxicillin (5th month – 10 days) | Complementary feeding |
| 12 | M | Amoxicillin (2nd and 9th months – 7 days) | Exclusive breastfeeding |
| 13 | M | NO | Exclusive breastfeeding |
| 14 | M | Amoxicillin (9th month – 10 days) | Exclusive breastfeeding |
| 15 | M | Cephalexin (10th month – 10 days) | Complementary feeding |
| 17 | M | Amoxicillin (6th month – 10 days) | Exclusive breastfeeding |
DNA was extracted from stools using Stool QIAmp DNA Mini kit (Qiagen®,CA, USA) according to the manufacturer's instructions. The extracted DNA from stool was quantified with a NanoDrop spectrophotometer (Thermo Scientific®, MA, USA) and kept at −20°C until use.
Polymerase chain reaction (PCR) amplification for DGGEThe PCR reaction for DGGE was performed using primers for the V3 hypervariable region of the 16S rRNA gene (F338GC: 5′-CGC CCG GC CCG GGC CGC GGC GGC GGG GGC CGG GGG GCA GGG GCC TAC GGG AGG CAG CAG-3′ and R518: 5′-ATT GCT GCT ACC GCG GG-3′).15 The reaction was optimized to a final volume of 25μL: 1X buffer, 1.5mM MgSO4, 0.2mM dNTPs, 0.2mM primers, 0.1U Hi-Fidelity Taq polymerase (Invitrogen®, CA, USA), and 20ng DNA. The reactions were performed using the program described by Muyzer et al.15
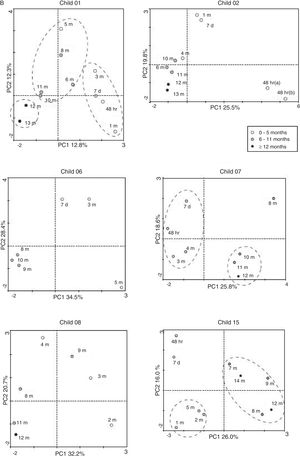
DGGE of PCR ampliconsPCR amplicons were separated by DGGE by using the specifications of Muyzer et al.15 and the Decode system (BioRad Laboratories®, CA, USA). Bionumerics v.5.1 (Applied Maths®, Belgium) was used for the evaluation of diversity profiles of each sample, calculation of the Dice coefficient, and cluster analysis. The analysis considered band presence/absence, the GC%-derived position of each band and the relative intensity of the bands to reflect the shared microbial community and the abundance of each population, respectively. The Bray–Curtis coefficient was used to create a similarity matrix among each sample.16 A cluster analysis was performed from the similarity matrix with Primer 6+.17 The correlation between samples and band profiles generated by DGGE was determined by correspondence analysis (CA) performed with the program Canoco v4.5 (Biometris®, Wageningen, Netherlands). The CA biplot was constructed with inter-species distance scaling, where each child was considered a distinct sample and each DGGE band was considered a unique species. No sample or species-weights were specified. In order to make data visualization clearer, three groups of DGGE profiles were created for each sample: (a) 0–5 months; (b) 6–11 months; (c) >12 months. Grouping circles were automatically drawn by the Canoco software for the three profile groups. In order to test whenever the type of diet influenced the microbial community, children were grouped into exclusive (child #12, 13, 14, and 17) and complementary (child #1, 2, 3, 6, 7, 8, and 15) breastfeeding. Child #3 was included in the complementary group for this analysis. A PERMANOVA analysis18 was done using the similarity matrix described above. A total of 999 permutations were done among DGGE profiles of children between the two diet groups. Since the design is unbalanced (different number of children in each group), a Type III sum of squares was used. The critical p-value was adjusted by Bonferri correction for multiple comparisons (alpha=0.05; adjusted p-critical=0.005).
Ethical considerationsThis research was approved by the Ethics Committee of the HU-USP (under registration number 574/05). All of the mothers enrolled in the research signed an informed consent.
ResultsClinical informationFrom the 11 children enrolled in this study, four had a diet characterized as exclusive breastfeeding up to the 6th month of age. One child had a diet characterized as predominant breastfeeding up to the 6th month of age, with the introduction of tea with honey at the 3rd month, and the other six children had a diet characterized as complementary feeding up to the 5th month of age (Table 1), with the introduction of non-human milk, fruits, and yogurt at the 2nd or 3rd months of age. At the 6th month of age, most of the children were eating soft food such as soup, fruits, fruit juice, and yogurt. At the end of the 12th month of life, all children were eating a diverse solid diet, which included meat, grains (rice, beans, peas, and lentils), wheat flour, fiber (greens), fruits, milk (formula and/or breast milk), and yogurt. Seven received antibiotic for respiratory infection during the study (Table 1).
Molecular analysis of infants’ microbiota by DGGEFecal DNA of each child was extracted and the variable V3 region of the 16S rRNA gene was amplified. Clustering analysis was performed for all time points of all children to search for some correlation (Fig. 1S – supplementary material). It was not possible to correlate any pattern of similarity with breastfeeding practice by analyzing the dendrogram. Therefore, each DGGE profile was subjected to clustering analysis and a dendrogram was obtained for each child (data not shown). The individual results revealed an increase in bands over the months, indicating an increase in microbiota diversity and complexity, and in inter-individual differences in DGGE profiles, without a unique pattern seen for all children.
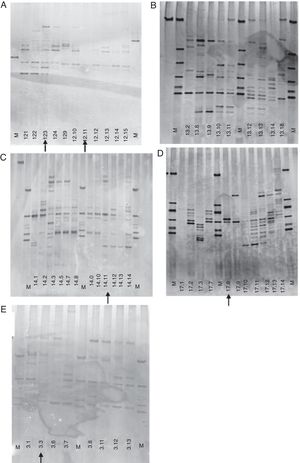
However, the PCR-DGGE profile of each child revealed an interesting pattern of similarity between time points for those who were on exclusive breastfeeding or predominant breastfeeding (Fig. 1). The microbiota profile of those children showed a persistence of prominent bands even after weaning and/or introduction of solid food, and also a gradual increase in the number of bands over time, suggesting an ecological succession. This was not observed for those children who had mixed feeding and introduction of solid food before the 5th month of life (Fig. 2), for whom a varied profile was observed, without similarities at the same time point. The PERMANOVA analysis comparing exclusive and complementary breastfeeding revealed significant differences between these groups (p=0.004; adjusted p-critical=0.005). According to this result, the microbial community of each breastfeeding group is different at a 95% confidence level.
The DGGE profile of children under exclusive breastfeeding showed a microbial profile characterized by a stable pattern of bands, with common bands during the study time, with a slight increase or decrease in intensity (Fig. 1A–D), even during the influence of external factors, such the introduction of solid food or antibiotic administration. In the first weeks of life, there were a few bands that disappeared in subsequent months, but then a pattern was established, with an increase in bands, indicating an increase in diversity. After solid food introduction into the diet, the band profile changed, with the disappearance of some bands, and the appearance of others, although the prominent bands remained present with no differences in intensity, suggesting an ecological succession, and an increase in diversity.
Children #12, 13, 14, and 17 were on exclusive breastfeeding up to the 6th month of age, and soft food and fruit juice were then introduced during this month. Only child #13 did not receive an antibiotic, while the others were given antibiotic once or twice to treat upper respiratory infections (Table 1). For children who received antibiotic, the diversity was reduced, and there was a fluctuation in the intensity of the bands in the month following drug administration. However, the microbial profile remained stable, and diversity increased in the subsequent months.
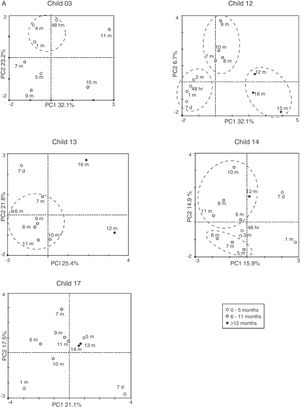
Although there were changes in intensity and fluctuation of some bands, the DGGE pattern in the one-year microbial analysis was stable for breastfeeding infants. There was ecological succession with food introduction or antibiotic administration, without losing the microbial community, suggesting microbiota resilience. This evolution was also reflected in the CA. An ecological succession was observed after the introduction of food for all of these infants; at the 6th month of age, with a shift in microbial profile, and after the 12th month of age, the microbial community seemed to be preserved (Fig. 3A).
Predominant breastfeedingThe DGGE profile in this diet was similar to one observed in exclusive breastfeed. Only one child (#3) had a diet characterized as predominant breastfeeding, with an introduction of tea with honey at the third month of age (Fig. 1E). This child had a skin infection at the 7th day of life10 and was treated with cephalexin for ten days. The DGGE profile showed a reduction in bands in the months following antibiotic administration, with a recovery of diversity at the 5th month of age, with an evident pattern in the microbial community. The introduction of soft food at the 6th month did not seem to disturb this pattern, and the community structure was observed up to the end of the study time. Even the introduction of tea did not affect this pattern. This profile was also noticed in the CA, which indicated the change in microbial community structure after the 5th month of age (Fig. 3A).
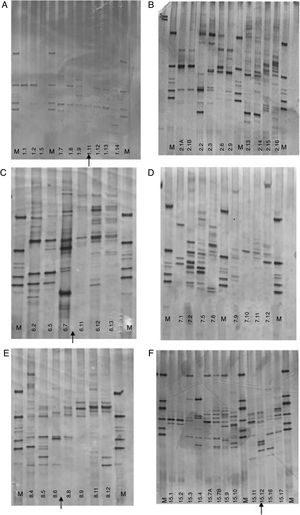
Complementary feedingThe DGGE profiles observed for the children not on exclusive breastfeeding (Fig. 2) showed a fluctuation in bands, without a restoration of the initial microbial pattern. They had a mixed feeding at the second month of life, and solid food was introduced at the 3rd or 4th month of age. The DGGE profile did not show a pattern of ecological succession as seen with the children described above, but rather a fluctuation in bands over the study time. There was an increase in bands over time in a bigger proportion of the profile compared to breastfeeding children; however, the microbial community structure was not preserved, and no pattern could be identified. The children under complementary food were #1, 2, 6, 7, 8, and 15. Only children #2 and 7 did not receive any antibiotics during the study time, and even so, the DGGE profiles were dispersed without a defined pattern. For those who received antibiotics, the number of the bands in the DGGE profile decreased during the antibiotic administration time (Fig. 2), and an increase in bands was observed in subsequent months, but with a different microbial community. CA showed a shift in microbial profile after the introduction of non-human milk or soft food before the 6th month of age, disturbing the microbial community (Fig. 3B).
DiscussionOur group had already described; using qPCR and 16S rRNA library construction, the composition of intestinal microbiota of this group of children in some points during their first year of life.5,10 In the present study, however, the microbial community structure was analyzed in a monthly basis, revealing a more detailed view of these changes over time.
The dendrogram analysis did not show any correlation between clustering profile and breastfeeding practice, since there were inter-individual differences between the children, as discussed previously.5 On the other hand, the analysis of the individual's profile allowed a clearer interpretation of intestinal microbial community structure, showing an increase in bands over the months, indicating an increase in microbiota diversity and complexity, as previously discussed.3,5,19
Bacterial resilience was described as the rate at which microbial composition returns to its original composition after being disturbed,20 due to physiological features and adaptation mechanisms in the new environment. If abundance was reduced by a disturbance, some bacteria group may benefit from the new conditions and then increase in abundance, restoring the original composition.20
The analysis of DGGE results of this group of children revealed an interesting profile of microbiota community resilience in children fed exclusively with breast milk, supported by the PERMANOVA analysis, in which the microbiota profile was different from those under complementary feeding (p<0.005). Children #12, 13, 14, and 17 were on exclusive breastfeeding up to the 6th month of life, and after the introduction of solid food and/or antibiotic administration, an increase in band number was noted, suggesting an increase in microbial diversity. Moreover, the DGGE patterns were not disturbed and a structure of the community was shared in each child, as seen by CA. Despite the impact on the diversity of bacterial genera, the introduction of solid food and/or antibiotic administration had little effect on bacterial community structure, showing evidence of microbiota resilience.
Breastfeeding is universally recognized as the ideal way to feed infants with a complete source of nourishment,21 and also it effectively reduces morbidity and mortality from diarrhea and other infectious diseases.22
Human milk is a source of symbiotic and probiotic bacteria23 and prebiotics24 for the baby's gut, such as human milk oligosaccharides (HMOs). HMOs are not digested by the human gut, and therefore, the role of intestinal microbiota is crucial for their hydrolysis.24 In this way, HMOs increase the population of beneficial bacteria by both probiotic and prebiotic effects, and, consequently, the intestinal microbiota of infants is dominated by Bifidobacterium and Lactobacillus.25,26
The introduction of solid food in children's diet provides a great change in intestinal microbiota diversity.13,27,28 At the end of the 6th month of age, all children had a diverse solid food diet, which included meat, grains (rice, beans, peas, and lentils), wheat flour, fiber (greens), fruits, milk (formula and/or breast milk), and yogurt.5 The DGGE patterns of the children studied showed an increase in bands, suggesting also an increase in microbiota diversity. In children with complementary feeding, the DGGE profile showed a fluctuation in bands during the time studied, and also no sharing of common bands according to CA, corroborating the DGGE results. Furthermore, the early introduction of solid food in the baby's diet seemed to be associated with instability in the microbiota community.
Child #3 had a diet characterized as predominant breastfeeding, with the introduction of tea with honey at the 3rd month of age and breast milk up to the 6th month of age. The DGGE pattern was similar to that of children on exclusive breastfeeding, and the tea and honey did not seem to disturb the microbial ecology over time. Despite the change in microbial richness,10 the administration of antibiotics did not affect microbial resilience, as evident also for children on exclusive breastfeeding.
Antibiotic administration is also another external factor that has a negative impact on intestinal microbiota diversity,5,29 with a perturbation of microbiota resilience.30 However, the three children (#12, 14, and 17) who were given oral antibiotics showed a reduction in bacterial diversity without disturbance of the intestinal microbial community. The microbial community appeared to be restored in the subsequent month after antibiotic administration.
The DGGE profile of child #14 demonstrated a marked decrease in microbial diversity after the introduction of solid food, with the restoration of diversity after the administration of antibiotics. However, the community structure was not altered, and the CA showed a pattern of shared bands without disturbances. Interestingly, the medical chart for this child showed a decline in growth curve after the 6th month, with signs of malnutrition over time. This child had less to eat due to socioeconomic factors, and the mother replaced food with breast milk, which could explain the decrease in microbial diversity and the maintenance of microbiota resilience.
The microbial profile of child #17 demonstrated a fluctuation in bands and appeared to give rise to a microbiota with a new profile, although it grouped with the other samples sharing the same bands, suggesting microbiota resilience. It is possible that this change was due to antibiotic administration concomitant with the introduction of solid foods. The effects of antibiotics on the microbiota did not allow new bacteria from foods or the prebiotic effects of foods to produce microbiota changes from the 6th to 10th month of life.
In the previous studies enrolling this group of Brazilian children,5,10,25 the authors showed a notable absence of Staphylococcus as intestinal colonizer10 and an abundance of E. coli,5 corroborating the worldwide knowledge of the importance of the influence of external factors in intestinal microbiota colonization. These children had a milk diet, and the intestinal microbiota was dominated by Bifidobacterium and Lactobacillus.25 However, none of these studies were able to associate specific profiles or changes in microbiota composition with exclusive breast feeding.
Using DGGE, the intestinal microbial community structure during the first year of life of the children studied was determined, and the results suggested the resilience of the intestinal microbiota pattern to external forces, here represented by antibiotic administration and the introduction of solid food. The maintenance of a stable bacterial community in the infants’ gut may provide eubiose effects in health over the following years. After birth and up to the weaning period, human milk is the only nutrition recommended by WHO.14 To the best of the authors’ knowledge, this is the first report showing the association of exclusive breastfeeding with microbial community resilience, reinforcing the importance of exclusive breastfeeding up to the 6th month of life, as recommended by the WHO.14
FundingFinancial support was from FAPESP (Fundação de Amparo à Pesquisa do Estado de São Paulo): No. 06/55141-4 to MBM and No. 11/51196-7 to CRT.
Conflicts of interestThe authors declare no conflicts of interest.
Dr. A. Leyva provided English editing of the manuscript.
Please cite this article as: Carvalho-Ramos II, Duarte RT, Brandt K, Martinez MB, Taddei CR. Breastfeeding increases microbial community resilience. J Pediatr (Rio J). 2018;94:258–67.
Study conducted at Universidade de São Paulo (USP), Faculdade de Ciências Farmacêuticas, São Paulo, SP, Brazil.