The aim of this study was to evaluate the association between possible functional interleukin-10 (IL-10) polymorphisms, IL-10 expression and regulatory T cells (Tregs) frequency, and/or asthma severity in a sample of children and adolescents.
MethodsThis is a nested case-control genetic association study. The study sample consisted of children and adolescents aged 8–14 from public schools. Four polymorphisms of the IL-10 gene (rs1518111, rs3024490, rs3024496, rs3024491) were genotyped in asthmatic subjects and controls using real-time PCR. Tregs cells and IL-10 were analyzed in peripheral blood mononuclear cells by flow cytometry. The severity of asthma was defined according to the Global Initiative for Asthma (GINA) guideline.
ResultsOne hundred twenty-three asthmatic subjects and fifty-eight controls participated in the study. The single nucleotide polymorphism (SNP) rs3024491 (T allele) showed association with asthma severity, presenting a higher frequency in patients in the moderate asthma group. The T allele of variant rs3024491 also showed an association with reduced IL-10 levels (p=0.01) and with increased Tregs frequency (p=0.01). The other variants did not present consistent associations.
ConclusionsOur results suggest that moderate asthma is associated with a higher frequency of the T allele in the SNP rs3024491. In addition, the variant rs3024491 (TT) was associated with a reduction in IL-10 production and an increased percentage of Tregs cells, suggesting possible mechanisms that influence asthma severity.
Asthma is a chronic, multifactorial, and polygenic disease caused by a complex combination of environmental stimuli, host factors, and genetic susceptibility. It affects more than 300 million people worldwide1 and is the most common chronic non-communicable disease in children.1,2 It is well-known that asthma often begins in childhood but can occur at all ages and at any time throughout life.1 Asthma presents higher incidence and prevalence in children, with morbidity and mortality rates higher in adults. Childhood asthma is more common in boys while adult asthma is more common in women.3
In the last few decades, a wide variety of environmental determinants and modern lifestyles have contributed to important changes in the epidemiology of asthma, such as an increase in environmental pollutants, obesity, and westernization. The International Study of Asthma and Allergies in Childhood (ISAAC) has been the most standardized methodology to provide worldwide comparable information on the epidemiology of asthma in children and adolescents. The population of interest in Phases One and Three was schoolchildren aged 6–7 years and adolescents 13–14 years. The ISAAC Phase Three research included 1.2 million children from 233 centers in 98 countries. This important study showed great variability in the prevalence of asthma between regions, continents, countries, and centers in the same country.2 The highest prevalence (≥20%) was generally observed in English speaking language countries, and in parts of Latin America.1,2 In Brazil, Phase Three of this study showed an average asthma prevalence of 24.3% (ranging from 16.5 to 31.2%) for children and 19% (11.8% and 30.5%) for adolescents, unrelated to socioeconomic status.4
In addition to environmental stimuli, genetic factors have a strong influence on the etiology of asthma. Studies with mono and dizygotic twins have shown that asthma has a heritability of approximately 60%.5,6 Moreover, inflammatory cytokines produced by Th2 cells play an important role in asthma. However, other subtypes of CD4+ T cells are also involved in its pathogenesis.7 Interleukin-10 (IL-10) is a pleiotropic cytokine with an important anti-inflammatory and immunoregulatory function, playing a protective role in asthma.8 Low levels of IL-10 have been associated with asthma.9 Also, IL-10 may potentiate the differentiation of regulatory T cells (Tregs).10 Some studies have shown changes in the number and function of Tregs in asthma.11–15
Single Nucleotide Polymorphisms (SNPs) have been related to asthma pathogenesis. The IL-10 gene is located at chromosome 1 (1q31-32), a region associated with asthma susceptibility. The SNPs of the IL-10 gene most studied are in the promoter region and comprise the variants rs1800896 (-1082A/G), rs1800871 (−819T/C) and rs1800872 (-592A/C). These SNPs show linkage disequilibrium (LD) with other polymorphisms of the gene and have been reported to have a regulatory role on IL-10 expression.16 Three meta-analysis studied these three polymorphisms in children and adults, but the results were inconclusive.17–19 A recent meta-analysis evaluated the association between IL-10 promoter polymorphisms and the risk of pediatric asthma. The results of this study indicated that the IL-10 -1082G/A polymorphism might be a risk factor for asthma in children.20
Thus, the objective of this study was to evaluate the association between possible functional IL-10 polymorphisms, IL-10 expression, Tregs cells frequency, and asthma severity in a sample of children and adolescents.
MethodsStudy design and population sampleThis is a nested case-control genetic association study. The study sample consisted of children and adolescents aged 8–14 from public schools. The study was stratified into three phases: 1. ISAAC questionnaire to assess the prevalence of asthma in schoolchildren; 2. Assessment of asthma control and severity through simple procedures such as Global Initiative for Asthma (GINA)21 questionnaires, and a pulmonary function test; 3. Blood collection for evaluation of atopy with specific IgE for analysis of inflammatory markers and for genotyping in a case-control sub-sample.
The ISAAC questionnaire was used to select the population of asthmatics. The control group included children from the same schools, without diagnosis of asthma (ISAAC questionnaire). Afterwards, the children diagnosed with asthma and selected controls underwent a series of evaluation procedures in phases 2 and 3: spirometry, tests for specific IgE and analyses of inflammatory markers. Finally, the samples were genotyped for IL-10 SNPs. The severity of asthma was defined according to the GINA guideline21 and the subjects were classified as having mild, moderate and severe asthma.
Exclusion criteria included cardiovascular disease, immunodeficiencies, chronic respiratory diseases, recent asthma/rhinitis exacerbation, acute respiratory infections or use of systemic steroids in the last four weeks.
Assessment of atopy and lung functionAtopy was defined by specific IgE (ImmunoCAP, Phadia AB, Uppsala, Sweden) for the aeroallergens (Dermatofagoides pteronyssinus, Dermatofagoides farinae, Dust and Blomia tropicalis, Periplaneta americana, dog and cat, fungi, pollen and grass). Children with a detection level of at least 0.35kU/L of any of the allergens tested were considered atopic.22
Trained professionals performed spirometry (Koko equipment, Ferraris Respiratory, Louisville, CO, USA) according to the ERS/ATS guidelines.23 At least three acceptable tests were obtained before and after the use of a bronchodilator (salbutamol, 400μg). The results of the variables forced expiratory volume in the 1st second (FEV1), forced vital capacity (FVC), FEV1/FVC ratio and forced expiratory flow between 25% and 75% (FEF 25% -75%) were described into z scores according to reference values of the Global Lung Function Initiative (GLI 2012), adjusted for height, age and sex.24
Selection of IL-10 polymorphismsIL-10 polymorphisms were initially selected by project database (version 28.0) in order to include the most frequent genetic variations.25 For the present study, we identified the SNPs rs1518111, rs3024490, rs3024496 and rs3024491, with minor allele frequency (MAF) of 42%, 40%, 31%, 28%, respectively.
Blood collection for gDNA extractionBlood samples were collected during visits at the outpatient clinic. Ten mL of blood was collected from each study participant. Aliquots of 1mL were stored in a freezer (-20°C) for later use in the extraction of genomic DNA (gDNA), following the technique of phenol/ chloroform/ isoamyl alcohol extraction (25: 24: 1), already described.26
GenotypingSample genotyping was performed using the Real-Time PCR (TaqMan-Applied Biosystems, Thermo Fisher Scientific, Waltham, MA, USA) following the manufacturer's instructions. 0.1ng or 0.25ng of gDNA were used for the real-time PCR reaction. The genotyping master mix, catalog 4371355 (Applied Biosystems, Thermo Fisher Scientific, Waltham, MA, USA) and the primers of IL-10, SNP genotyping assays rs1518111, rs3024490, rs3024496 and rs3024491 (Applied Biosystems, Thermo Fisher Scientific, Waltham, MA, USA) were used for genotyping with the support of the StepOne ™ Real-Time PCR System (Applied Biosystems, Thermo Fisher Scientific, Waltham, MA, USA). The data obtained from the amplification (threshold cycle-CT) were analyzed by the StepOne program (version 2.3). The result was expressed as a frequent homozygous, heterozygous, rare homozygous or indeterminate.
Analysis of regulatory T cell frequency and IL-10 productionFor analysis of regulatory T cell frequency, peripheral blood mononuclear cells were purified from 9mL of whole blood using Histopaque-1077 separation gradient (Sigma-Aldrich Corporation, St Louis, MO, USA) and frozen at −80°C. After, 2×105 cells were labeled with anti-CD4 PE Cy7 (clone SK3-BD Biosciences, Franklin Lakes, NJ, USA) and anti-CD25 APC H7 antibodies (clone M-A251-BD Biosciences, Franklin Lakes, NJ, USA) for 30min. Cells were permeabilized using FoxP3 buffer (BD Biosciences, Franklin Lakes, NJ, USA) and then labeled with anti-FoxP3 Alexa Fluor®488 antibody (clone 259d/ C7, BD Biosciences, Franklin Lakes, NJ, USA) for 20min at room temperature. The cells were analyzed by flow cytometry (FACS Canto II, BD Biosciences, Franklin Lakes, NJ, USA), and data were analyzed with FlowJo software (Tree Star, Ashland, OR, USA). Isotype control antibodies were used to exclude non-labeled cells specifically. For analysis of IL-10 production, peripheral blood mononuclear cells were purified and cultured with anti-CD3 and anti-CD28 antibody for 24h, and the supernatant was collected and analyzed using CBA (BD Bioscience, Franklin Lakes, NJ, USA) by flow cytometry.
Statistical analysisThe quantitative variables were evaluated using the Kolmogorov-Smirnov test. Data that presented normal distribution were presented in mean and standard deviation and asymmetric data, in median and interquartile range. The qualitative variables were expressed in absolute and relative frequency. The comparison between the quantitative data was performed by Student's t-test for independent samples or by the Mann-Whitney U test The categorical variables were compared using the Pearson Chi-Square Test. For analysis of Tregs cells frequency and IL-10 production, the GraphPad Prism 5 program (San Diego, CA, USA) was used. The other analyses and data processing were performed with SPSS version 18.0 (SPSS Inc., USA). Differences were considered significant when p<0.05.
Ethical aspectsAll guardians of the children and participants signed the informed consent. The identity of all the participants was kept confidential. First, the project was approved by the University Scientific Board (number 6.139). Then, the study was registered at the Brazilian Research Database called Plataforma Brasil - CAAE number 19835913.8.0000.5336 (http://plataformabrasil.saude.gov.br), and approved by the Human Ethics Committee of the Institution, under the number 10/04978.
ResultsIn this genetic association study, 123 cases and 58 controls were included and genotyped (N=181). There was no significant difference between the groups regarding the variables age, sex and birth weight (Table 1). Patients with asthma presented atopy more frequently (75.6% vs 54.4%, p=0.004) and reduction in some lung function, such as FEV1/FVC (-0.50±0.91 vs -0.13±0.70, p=0.013) and FEF25% -75% (-0.43±1.06 vs -0.00±0.91, p=0.015). Patients with asthma also had greater frequency of the FoxP3 in CD4+ and CD25+ T cells, presenting a CD4+CD25+Foxp3+ median frequency of 1.38 (0.39–3.25) compared to controls 0.25 (0.02−0.52); p<0.0001 (Table 1).
Descriptive data of the sample comparing the study group versus the control group.
| Variables | Asthma(n=123) | Controls(n=58) | p |
|---|---|---|---|
| Age, years | 11.17±1.09 | 11.28±1.24 | 0.537 |
| Male gender, n (%) | 63 (51.2) | 27 (46.6) | 0.558 |
| Birth weight, g | 3.40±0.57 | 3.29±0.45 | 0.228 |
| Atopy, n (%) | 90 (75.6) | 31 (54.4) | 0.004a |
| FEV1/FVC, Z score | −0.50±0.91 | −0.13±0.70 | 0.013b |
| FEF25−75%, Z score | −0.43±1.06 | −0.00±0.91 | 0.015b |
| CD4+CD25+FoxP3+,(% of cells) | 1.38 (0.39–3.25) | 0.25 (0.02 – 0.52) | < 0.0001c |
FEV1, forced expiratory volume in the 1st second; FVC, forced vital capacity; FEV1/FVC, Tiffenou index; FEF (25−75%), forced expiratory flow between 25 and 75%; CD4+CD25+FoxP3+, Treg cells.
Four common IL-10 SNPs (rs1518111, rs3024490, rs3024496 and rs3024491) were selected for genotyping. The frequencies of the rare alleles for each SNP were 38.6%; (A allele, rs1518111), 40.6% (G allele, rs3024490), 29.8%, (T allele, rs3024496), 27.4% (G allele, rs3024491) (Table 2). The selected SNPs are in LD with other polymorphisms of the gene and represent a large part of the loci variations.
IL-10 gene variants included in the study.
| SNP | Position on the chromosome | Gene location | Allele | MAF (HapMap) | MAF (Sample) |
|---|---|---|---|---|---|
| rs1518111 | 206944645 | Intron 1 | A/G | 0.42 | 38.62 % (A) |
| rs3024490 | 206945311 | Intron 1 | G/T | 0.40 | 40.65% (G) |
| rs3024496 | 206941864 | Exon 5 | C/T | 0.31 | 29.83% (T) |
| rs3024491 | 206771701 | Intron 2 | G/T | 0.28 | 27.45% (G) |
HapMap, Haplotype Map; MAF, Minor Allele Frequency; SNP, Single Nucleotide Polymorphism.
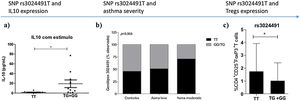
The IL-10 SNP rs3024491 showed a trend for association with disease severity when assessed for frequency in patients with moderate asthma, mild asthma and controls (Fig. 1b). However, when we analyzed mild asthmatic patients and controls versus moderate asthma, IL-10 rs3024491 SNP showed a significant association with asthma severity, presenting a higher frequency of T allele in patients in the moderate asthma group versus mild asthma and controls (71. 4% vs. 48.5%, chi-square test; p=0.042).
In the analysis with a dominant model, TT genotype (rs3024491) was shown to be associated with a reduction in IL-10 levels; p=0.01 (Fig. 1a). In this same model, the TT genotype of the variant rs3024491 was shown to be more frequent in patients with moderate asthma than in controls and patients with mild asthma (Fig. 1b). Finally, the same rs3024491 TT genotype also demonstrated association with increased frequency of regulatory T cells; p=0.01 (Fig. 1c). The variant rs3024496 showed an association only with increased frequency of Tregs cells (p=0.01) in the C allele. The other variants of the IL-10 gene, rs1518111 and rs3024490, were not significantly associated with increased Tregs cells.
DiscussionIn this study, the rs3024491 variant of the IL-10 gene showed the most consistent and significant findings. The T allele of variant rs3024491 of the IL-10 gene showed association with asthma severity and was associated with lower expression of IL-10. This polymorphism (rs3024491) may influence the IL-10 expression or function. In addition to the association with asthma severity (Fig. 1b), this polymorphism also demonstrated to influence Tregs cell frequency (Fig. 1c).
Our study also showed that children with asthma had a higher percentage of Tregs cells in peripheral blood than controls; p<0.0001 (Table 1). In accordance with our findings, a few studies observed an increased frequency of Tregs cells in asthmatics compared to controls.11–13 Raedler et al. also found increased Tregs cells in atopic asthmatics compared to healthy controls. This increase of Tregs cells in asthmatic patients is probably due to a mechanism of counterbalance, wherein an inflamed environment, the immune system reacts producing more Tregs to counterbalance or contain the inflammation.12 On the other hand, other studies show lower percentage of Tregs cells in asthmatic children.14,15 Further studies are needed to confirm this hypothesis.
IL-10 polymorphisms have been extensively studied in several inflammatory and auto-immune diseases. Associations of these three variants of the IL-10 gene with rheumatoid arthritis,27 systemic lupus erythematosus,28 Crohn's disease,29 and psoriasis30 have been described. Some studies have identified an association between IL-10 gene polymorphisms and asthma risk, but the results were inconclusive. Three previous meta-analyses analyzed polymorphisms in the IL-10 gene promoter region (-1082/ -592/-819) and the risk of asthma.17–19 Nie et al. evaluated 18 case-control studies (4478 asthmatics and 4803 controls) and found significant associations between the -1082A/G and -592A/C polymorphisms of the IL-10 gene and the risk of asthma in Asians and Caucasians. Individuals AA homozygosity (-1082A/G, in LD with rs3024491) had a 27% increase in disease risk. Carriers of the A allele in the -592A/C polymorphism also had an increased risk of asthma.17 Hyun et al. evaluated 11 studies (2215 asthmatics and 2170 controls). Three of these studies were conducted in the European population and eight in the Asian population. Four studies were conducted only with children and an additional four with children and adults. This meta-analysis also demonstrated that the -1082G/A polymorphism confer susceptibility to asthma. The polymorphisms -592C/A, and -819C/T were not associated with the disease.18 In the meta-analysis of Zheng et al., 23 studies (4716 asthmatics and 5093 controls) were evaluated. In the 23 studies, Asian, Caucasian and African population samples were analyzed. This meta-analysis found that the promoter polymorphisms -1028A/G and -592A/C are probably associated with asthma susceptibility, especially in Asian samples with atopic asthma.19
Interestingly, there are high levels of LD between the promoter polymorphisms and the variants evaluated in our study (especially -1082 A and rs3024491T). The IL-10 gene is highly polymorphic and several variations within it or close to it may influence the IL-10 expression.31 The variant rs3024491 was shown to be approximately 100% in LD with the promoter region variant, rs1800896 (-1082A/G).32 Therefore, rare alleles may have the same functional consequences measured by the outcomes. Thus, one may suspect that only one of them causes the final effect, but the association between SNPs and asthma is probably the same. Most genetic association studies were performed with IL-10 promoter variants. In our study, we used four SNPs of the IL-10 gene that had high minor allele frequency (MAF) and were distributed along the gene structure. Some of them (especially rs3024491) are in LD with the IL-10 promoter SNPs.
This study presented some limitations and sample size is probably the most important. However, the consistency of the association results found at different levels (genetics, IL-10 expression, Tregs and phenotype) demonstrate the strength and possible impact of the result, despite the small sample for a genetic association study. Another limitation is that we did not have patients with severe asthma and severe therapy-resistant asthma in the samples. A further possible limitation is that the Tregs cell analysis was performed only on peripheral blood. Ideally, biological samples should be taken directly at the site of inflammation, in bronchoalveolar lavage (BAL) or through bronchial biopsy. However, this type of sample requires an invasive procedure and has been often denied by ethics committees.
In conclusion, IL-10 genetic polymorphisms may be one of the mechanisms related to the variation of the severity of asthma. Its association with asthma outcomes may be of great clinical relevance and may be a specific target for therapeutic strategies. Our results suggest that moderate asthma is associated with a higher frequency rs3024491T. We showed that the rs3024491T is associated with a reduction in IL-10 production, a high frequency of moderate asthma and a consequent increase in Tregs cells frequency. However, there is a need for more genetic association studies in different population samples.
FundingThis study was financed by the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior – Brazil (CAPES) – Finance Code 001.
Conflicts of interestThe authors declare no conflicts of interest.
The authors would like to acknowledge the financial support of the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior – Brazil (CAPES) – Finance Code 001.