to determine whether C. trachomatis was present in neonates with infection, but without an isolated pathogen, who died during the first week of life.
Methodsearly neonatal death cases whose causes of death had been previously adjudicated by the institutional mortality committee were randomly selected. End-point and real-time polymerase chain reaction of the C. trachomatis omp1 gene was used to blindly identify the presence of chlamydial DNA in the paraffinized samples of five organs (from authorized autopsies) of each of the dead neonates. Additionally, differential diagnoses were conducted by amplifying a fragment of the 16S rRNA of Mycoplasma spp.
Resultsin five cases (35.7%), C. trachomatis DNA was found in one or more organs. Severe neonatal infection was present in three cases; one of them corresponded to genotype D of C. trachomatis. Interestingly, another case fulfilled the same criteria but had a positive polymerase chain reaction for Mycoplasma hominis, a pathogen known to produce sepsis in newborns.
Conclusionthe use of molecular biology techniques in these cases of early infant mortality demonstrated that C. trachomatis could play a role in the development of severe infection and in early neonatal death, similarly to that observed with Mycoplasma hominis. Further study is required to determine the pathogenesis of this perinatal infection.
determinar se a C. trachomatis está presente em neonatos com infecção, porém sem patógeno isolado, que morreram durante a primeira semana de vida.
Métodoscasos de óbito neonatal precoce cujas causas de óbito haviam sido anteriormente determinadas pelo Comitê de Mortalidade da instituição foram aleatoriamente selecionados. Foram utilizadas as reações em cadeia da polimerase convencional e em tempo real do gene omp1da C. trachomatis, para identificar, às cegas, a presença de DNA de clamídia nas amostras desparafinizadas de cinco órgãos (de autópsias autorizadas) de cada um dos neonatos mortos. Além disso, foram realizados diagnósticos diferenciais por amplificação de um fragmento do rRNA 16S de Mycoplasma ssp.
Resultadosem cinco casos (35,7%) a presença de DNA de C. trachomatis foi detectada em um ou mais órgãos. Havia infecção neonatal grave em três casos; um deles correspondente ao genótipo D de C. trachomatis. Curiosamente, outro caso preencheu os mesmos critérios, porém possuía uma reação em cadeia da polimerase positiva para Mycoplasma hominis, um patógeno conhecido por causar sepse em recém-nascidos.
Conclusãoa utilização de técnicas de biologia molecular nos casos de mortalidade infantil precoce mostrou que a C. trachomatis poderia desempenhar um papel no desenvolvimento de infecção grave e no óbito neonatal precoce semelhante ao observado com a Mycoplasma hominis. São necessários estudos adicionais para determinar a patogênese dessa infecção perinatal.
Neonatal mortality has been defined as mortality occurring during the first 28 days of life. It is mostly associated to infections that are either congenital or acquired after birth. These infections strongly impact the causes of death and abortion in non-industrialized countries.1,2Chlamydia trachomatis is the cause of the most sexually transmitted bacterial infection worldwide.3 The prevalence C. trachomatis infection during pregnancy is variable: in the United States, it is between 2% and 13.7%; in Brazil, between 2.7% and 10%,4 and in Mexico, between 4% and 28%.5 It is related to premature rupture of membranes, chorioamnionitis, premature birth, and the development of neonatal ophthalmitis and pneumonitis. It has also been related to high rates of low birth weight and perinatal mortality.4 Variable clinical manifestations have been observed, including staccato cough, prodromal rhinorrhea, and history of conjunctivitis, tachypnea, and fever. The most frequent findings in chest radiographies are interstitial infiltration of bilateral lung fields, hyperinflation, and atelectasis.6
The risk for vertical transmission of C. trachomatis is between 60% and 70%, and occurs during the infant's passage through the birth canal; however, there is some evidence that vertical transmission can also occur in utero, since newborns delivered by cesarean sections have also been born infected and with intact membranes.6–9
Recently, Rours et al.10 demonstrated the presence of chlamydial DNA in the placenta of pre-term products, and found an association of the DNA with the degree and progress of tissue inflammation. Currently, there is no good experimental model available for the effects of Chlamydia infections during pregnancy and its association with neonatal death.
Aside from the pulmonary and conjunctive tissues of newborns, Chlamydia has also been found in intestinal, genitourinary, myocardial, and nervous system tissues.11–13 The presence of Chlamydia in these other tissues suggests that it has an invasive capacity. The present study aimed to detect chlamydial DNA in different tissues of neonates who were diagnosed with “infection without an isolated pathogen” and died during the first week of life. Neonates whose evident cause of death was not infectious were also studied. Additionally, it was sought to identify the Chlamydia genotypes involved in these neonates who developed early infection.
MethodsEthics statementThis study's protocol was reviewed and approved by the institutional Ethics Committee of the School Medicine at the National Polytechnic Institute, in Mexico City, Mexico.
DefinitionsInfectionSevere neonatal infection was established by: a) finding of C. trachomatis omp1 gene fragments in two or more different organs; b) clinical and laboratory data consistent with infection in the neonate during his lifetime; c) mother with antecedents of infection risk; and d) histopathological diagnosis of placental chorioamnionitis and pneumonitis in the cadaver.
Preterm rupture of membranes (PROM)Rupture of the fetal membranes prior to the onset of labor, regardless of gestational age.14
Early neonatal deathDeath occurring before the seventh day of extrauterine life.15
Inclusion criteriaThe inclusion criteria were: availability of all selected organs, samples, and reports; updated analysis by the perinatal mortality committee with final dictum; and complete records.
Selection of autopsy samplesA random selection of 20% of the cases of early neonatal deaths that occurred between January 1 and December 31, 2003 and fulfilled the inclusion criteria was performed. Cases were separated into two groups according to the judgment or final diagnosis of the institution's perinatal mortality committee. Group 1 consisted of cases of death due to systemic infection16 without pathogen identification. Group 2 was formed by cases of death due to another cause that was not related with infection.
The autopsy organs, specifically lungs, kidneys, brain, liver, and bronchi, which were embedded in paraffin blocks for all participating cases, were located. The study included only cases that had all these organs available, as well as complete maternal and neonatal clinical records and histopathological studies of the autopsy and placenta. The pathologist in chief blindly restudied the histologic samples from the cadavers.
Placentas are usually routinely analyzed: hematoxylin and eosin–stained histologic sections are investigated for deciduitis, vasculitis, endometritis, or chorioamnionitis without special studies. Later, the entire material is discarded. For this reason, only the results of the single study of the placenta were included.
The rest of the plates was deparaffinized and processed for DNA extraction. All tests were performed in a blinded manner. Clinical and demographic data were obtained from hospital records.
DNA extractionTissue samples were deparaffinized with xylene at room temperature and washed with ethanol. The obtained tissue was then treated with proteinase K, and the DNA was obtained by the phenol-chloroform-isoamyl technique and ethanol precipitation as previously described.17
Chlamydia trachomatis detectionEnd-point polymerase chain reaction and real-time polymerase chain reaction were performed using PTC-100 system (MJ Research, Inc. - Waltham, MA, United States) and StepOne (Applied Biosystems - Carlsbad, CA, United States), respectively. The primers used for end-point polymerase chain reaction are those proposed by Dutilh et al.18 and amplify a fragment of the omp1 gene of C. trachomatis (5’-GCCGCTTTGAGT TCTGCTTCCTC-3’; 5’-CCAAGTGGTGCA AGGATCGCA-3’).
Each end-point polymerase chain reaction contained 1.75 mM MgCl2, 0.2 mM dNTPs, 25 pM of each proposed primer, 2.5 U Taq polymerase (GoTaq® Flexi DNA polymerase Promega© - Madison, WI, United States), and 5μL of the DNA sample, for a final volume of 25μL. The reaction mixture was incubated for 5min at 95°C, followed by 35 cycles of 1min at 95°C for denaturation, 1min at 59°C for alignment, 1min at 70°C for extension, and a final elongation step of 5min at 70°C. The sample was considered positive when an amplification product of 129 bp was obtained.
The real-time polymerase chain reaction was performed using 3 mM of 2.7 mM MgCl2, 25 pM of each of the proposed primers, 2.5 U Taq polymerase (Applied Biosystems), and 5μL of the DNA sample, for a final volume of 25μL. The primers used were designed by Primer Express 3.0 Sequence program (Applied Biosystems), the amplified region is inside the same amplicon of 129 bp obtained with Dutilh's primers Fw: 5’-CCTGCTGAACCAAGC CTTATG-3’ and Rv: 5’-AGGATCTC CGCCGAAACC-3’; probe: 5’-TCGACGGAATTC TGT-3’. The reaction mixture was preheated at 95°C for 20 s, followed by 40 cycles of 30 s at 95°C, 20 s at 60°C, and 20 s at 72°C.
Amplification and detection of Mycoplasma spp. for differential diagnosesEnd-point polymerase chain reaction was performed according to the protocol proposed by van Kuppeveld et al.19 The primers used were: MGSO: 5’-GCACCATCTGTCACTCTGTTA ACCTC-3’ and GPO-1: 5’-ACTCCTACGGGAGGCAGCAGTA-3’. Each polymerase chain reaction contained 20 pM/μL primers, 10 uM/μL dNTPs, 2 mM/μL MgCl2, 5 U/μL Taq polymerase, 3μL DNA, and 43μL water, for a final volume of 50μL. The reaction mixture was incubated at 94°C for 5min, followed by 35 cycles of denaturing at 94°C for 1min, alignment at 60°C for 1min, extension at 72°C for 1min, and a final extension at 72°C for 5min. A sample was considered positive if a 715 bp product was amplified.
To detect the species, the following primers were used with the same polymerase chain reaction protocol: Mychomp (5’-ATACATGCATGTCGAGCGAG-3’) and Mychomn (5’-CATCTT TTAGTGGCGCCTTAC-3’). Additionally, M. hominis was detected according to Grau et al.20 and U. urealyticum was detected according to Blanchard et al.21 with the primers U5 (5’-CAATCT GCTCGTGAAGTATTAC-3’) and U4 (5’-ACGACGTC CATAAGCAACT-3’).
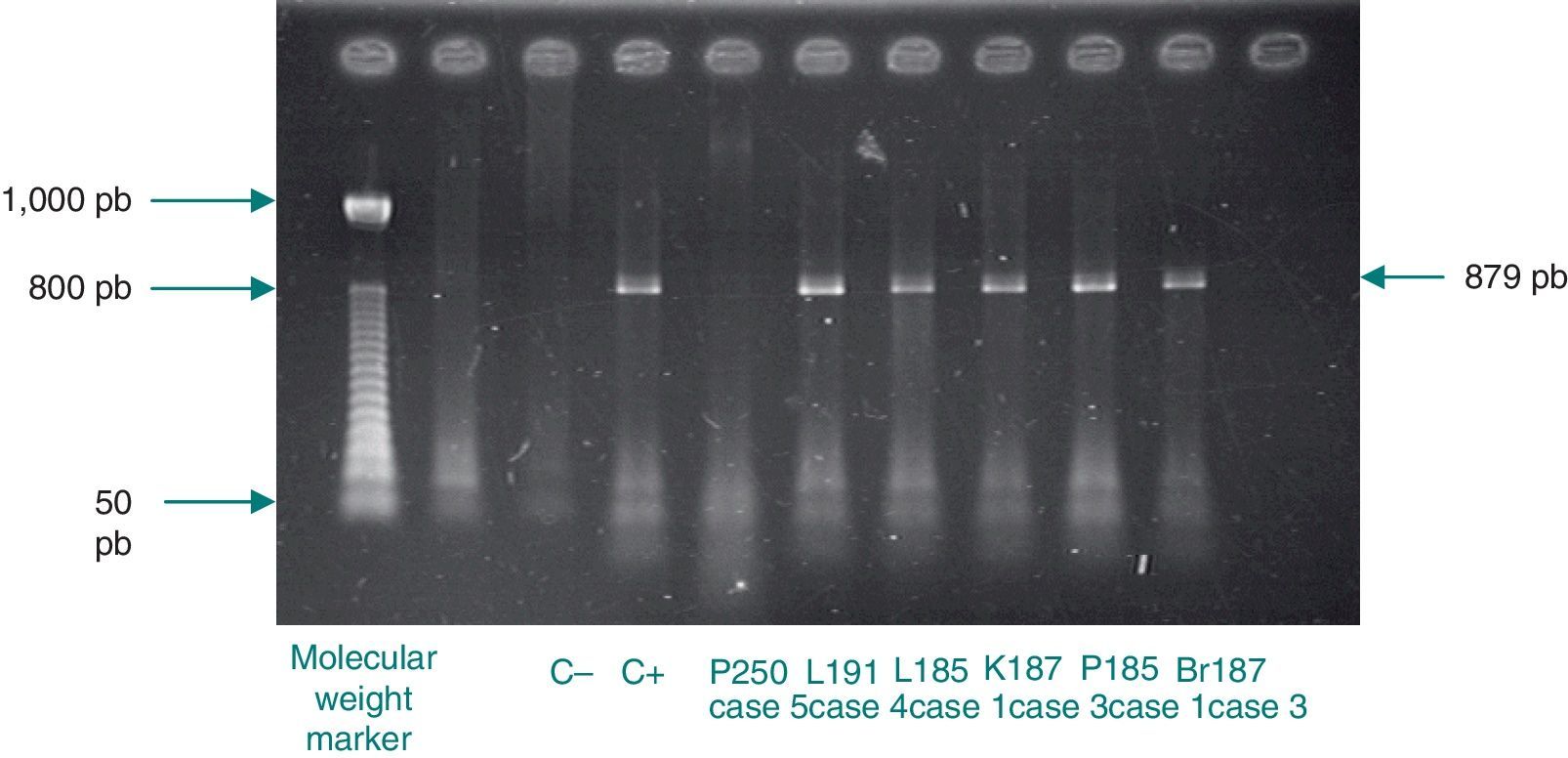
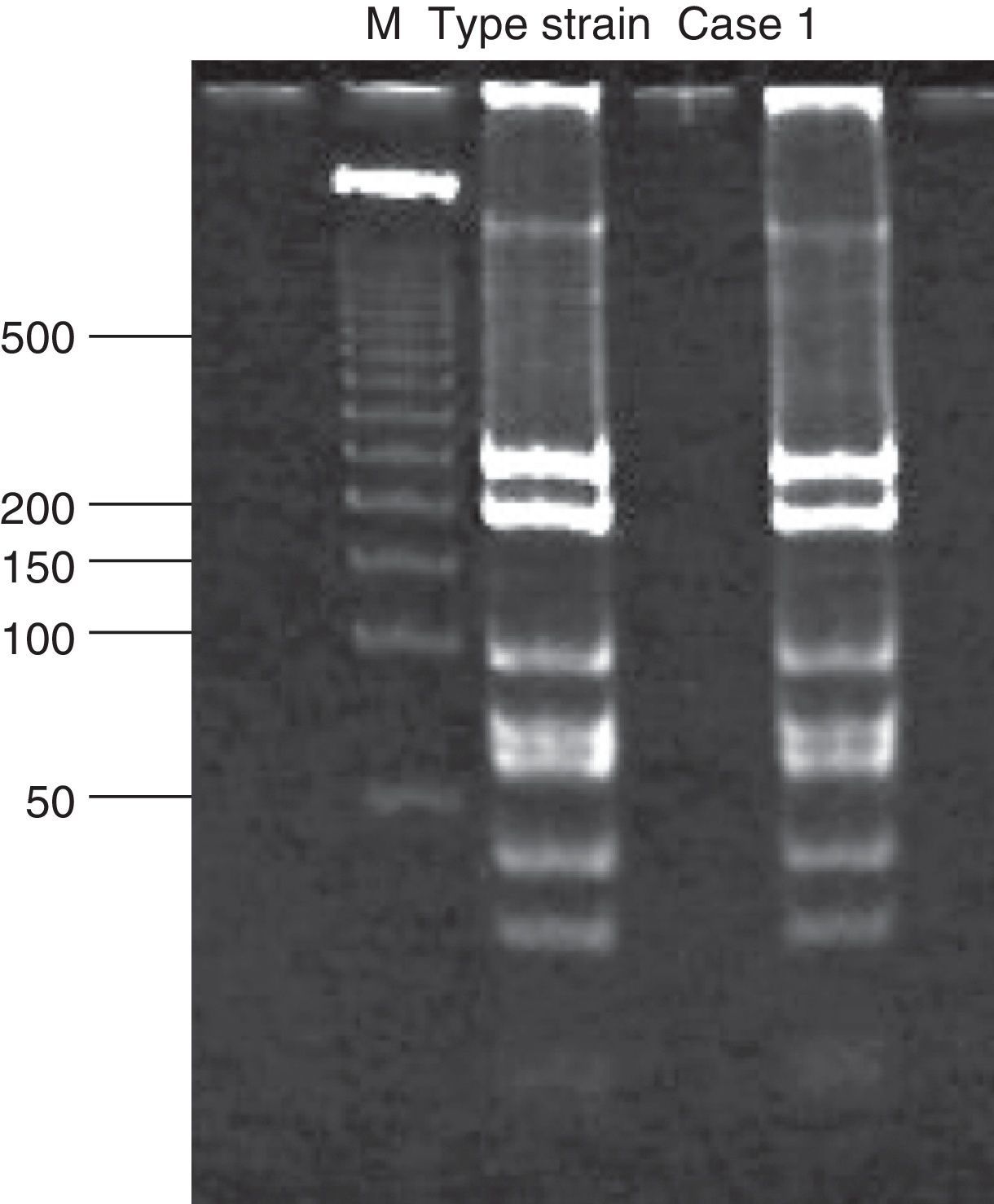
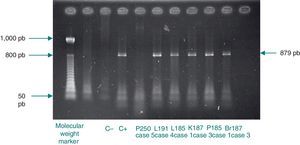
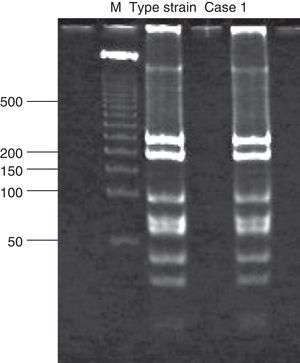
Analysis of restriction fragment length polymorphismA restriction fragment length polymorphism (RFLP) analysis was performed using a previously described method5 to genotype the samples with 129 bp amplicons. Briefly, a 1,142 bp fragment of the C. trachomatis omp1gene was amplified. The primers used for the amplification were the same as those reported by Yang et al.:22 OMP1 (5’-GCCGCTTTG AGTTCTGCTTCCTC-3’) and OMP2 (5’-ATTTACGTGAGCAGCTCTCTCAT-3’). Samples positive for the 1,142 bp amplicon were subjected to a second polymerase chain reaction that generated an 879 bp fragment. The primers used were also described by Yang et al.:22 P3 (5’-TGACTTTGTTTTCGA CCGTGTTTT-3’) and P4 (5’-TTTTCTAGATTTCATCTTGTTCAAT/CTG-3’). The 879 bp fragment was then incubated with the endonuclease Alu1 (Invitrogen, Applied Biosystems) for 10 h at 37°C. The resulting band pattern was compared with that of the corresponding type strain. In this study, that was uW-3Cx (ATCC VR-885D).
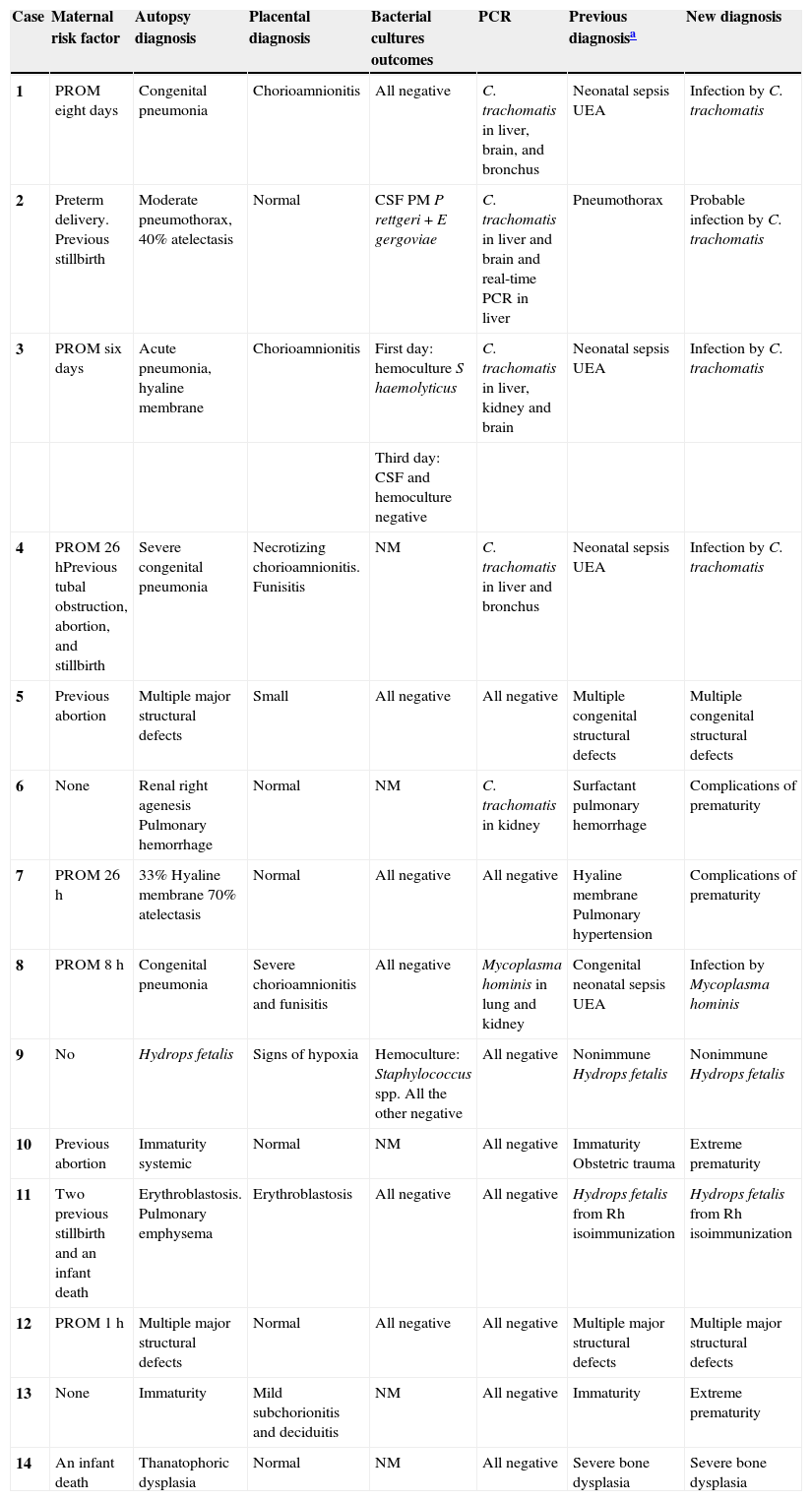
ResultsOf the 73 cases available, 18 were chosen at random. However, it was not possible to locate all the organs of interest in three cases, and the placenta study results were not available for one case. Table 1 depicts results of the 14 infant mortality cases studied, the perinatal characteristics of the newborns, and the most relevant maternal data. Four cases fulfilled all of the infection or neonatal systemic infection criteria (cases 1, 3, 4, and 8). Cases 1 and 8 were negative for bacterial and fungal cultures performed both during their life or postmortem. On the first day of life for case 3, Staphylococcus haemolyticus was isolated through blood culture. In case 4, the newborn lived only one hour, and the corresponding microbiological cultures were not performed.
Clinical and pathological correlations and outcomes of Chlamydia trachomatis (omp1) PCR in deparaffinized samples of brain, liver, kidney, lung, and bronchus. Early neonatal deaths.
| Case | Maternal risk factor | Autopsy diagnosis | Placental diagnosis | Bacterial cultures outcomes | PCR | Previous diagnosisa | New diagnosis |
|---|---|---|---|---|---|---|---|
| 1 | PROM eight days | Congenital pneumonia | Chorioamnionitis | All negative | C. trachomatis in liver, brain, and bronchus | Neonatal sepsis UEA | Infection by C. trachomatis |
| 2 | Preterm delivery. Previous stillbirth | Moderate pneumothorax, 40% atelectasis | Normal | CSF PM P rettgeri + E gergoviae | C. trachomatis in liver and brain and real-time PCR in liver | Pneumothorax | Probable infection by C. trachomatis |
| 3 | PROM six days | Acute pneumonia, hyaline membrane | Chorioamnionitis | First day: hemoculture S haemolyticus | C. trachomatis in liver, kidney and brain | Neonatal sepsis UEA | Infection by C. trachomatis |
| Third day: CSF and hemoculture negative | |||||||
| 4 | PROM 26 hPrevious tubal obstruction, abortion, and stillbirth | Severe congenital pneumonia | Necrotizing chorioamnionitis. Funisitis | NM | C. trachomatis in liver and bronchus | Neonatal sepsis UEA | Infection by C. trachomatis |
| 5 | Previous abortion | Multiple major structural defects | Small | All negative | All negative | Multiple congenital structural defects | Multiple congenital structural defects |
| 6 | None | Renal right agenesis Pulmonary hemorrhage | Normal | NM | C. trachomatis in kidney | Surfactant pulmonary hemorrhage | Complications of prematurity |
| 7 | PROM 26 h | 33% Hyaline membrane 70% atelectasis | Normal | All negative | All negative | Hyaline membrane Pulmonary hypertension | Complications of prematurity |
| 8 | PROM 8 h | Congenital pneumonia | Severe chorioamnionitis and funisitis | All negative | Mycoplasma hominis in lung and kidney | Congenital neonatal sepsis UEA | Infection by Mycoplasma hominis |
| 9 | No | Hydrops fetalis | Signs of hypoxia | Hemoculture: Staphylococcus spp. All the other negative | All negative | Nonimmune Hydrops fetalis | Nonimmune Hydrops fetalis |
| 10 | Previous abortion | Immaturity systemic | Normal | NM | All negative | Immaturity Obstetric trauma | Extreme prematurity |
| 11 | Two previous stillbirth and an infant death | Erythroblastosis. Pulmonary emphysema | Erythroblastosis | All negative | All negative | Hydrops fetalis from Rh isoimmunization | Hydrops fetalis from Rh isoimmunization |
| 12 | PROM 1 h | Multiple major structural defects | Normal | All negative | All negative | Multiple major structural defects | Multiple major structural defects |
| 13 | None | Immaturity | Mild subchorionitis and deciduitis | NM | All negative | Immaturity | Extreme prematurity |
| 14 | An infant death | Thanatophoric dysplasia | Normal | NM | All negative | Severe bone dysplasia | Severe bone dysplasia |
CSF, Cerebrospinal fluid; CT, Chlamydia trachomatis; NM, not made; PCR, polymerase chain reaction; PM, post-mortem; PROM, Preterm Rupture of Membranes (18); UEA, unknown etiologic agent.
The intentional search for DNA from Mycoplasma and Chlamydia in postmortem tissues using polymerase chain reaction resulted in amplification of the 129 bp C. trachomatis omp1 gene in five neonates. It was amplified in two or more organs for cases 1, 2, 3, and 4, and only in the kidney for case 6. In case 8, a 715 bp product, corresponding to Mycoplasma, was amplified in lung and kidney tissues (image not shown).
Chlamydial DNA was found in tissues of cases 2 and 6, but with no clinical or histopathological correlation indicating infection. These cases only presented with prematurity, barotrauma, and pulmonary hemorrhage. Additionally, amplification of 1,142 bp and 879 bp fragments for RFLP analysis of samples positive for chlamydial DNA was only achieved for cases 1, 3, and 4 (Fig. 1). The RFLP analysis of the samples where amplification of the 879 bp fragment was achieved only allowed for the identification of genotype D of C. trachomatis (case 1; Fig. 2). In cases 2 and 6, it was not possible to amplify the 1,142 bp fragment, and the presence of chlamydial DNA was confirmed in liver tissue through real-time polymerase chain reaction only for case 2 (image not shown).
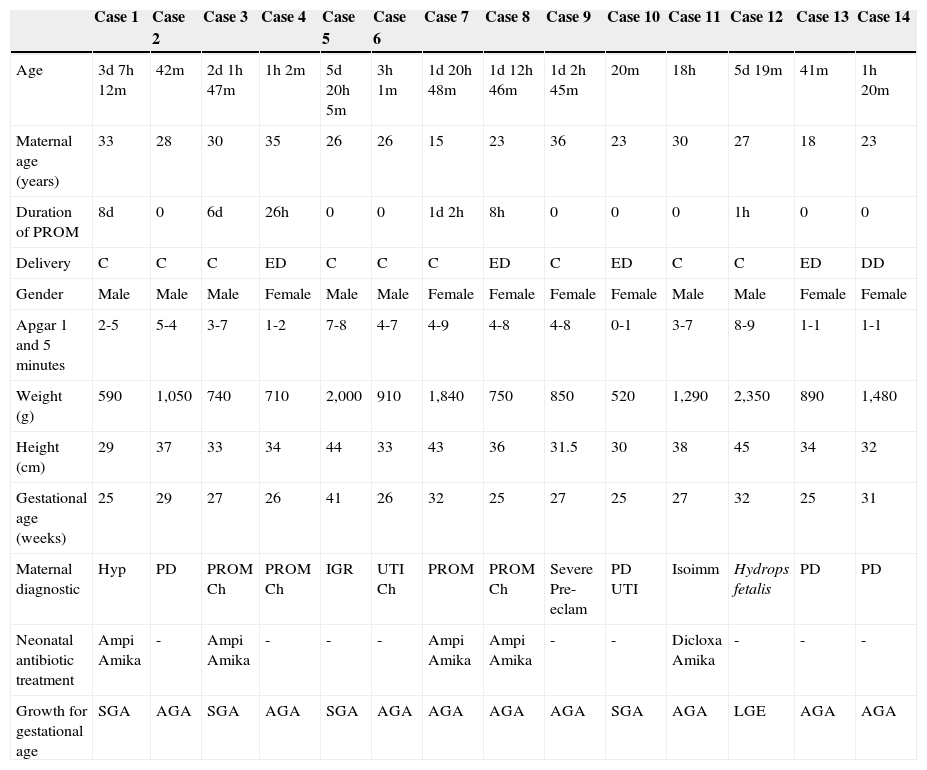
All cases were endotracheally intubated and received ventilatory assistance during their entire life. Six cases of premature membrane rupture were found. Of these, two neonates (cases 1 and 3) had evidence of chlamydial DNA and remained in utero with ruptured membranes for 8 and 6 days, respectively. The clinical characteristics of all cases are shown in Table 2.
Clinical characteristics (mother and child) of the infant mortality cases.
| Case 1 | Case 2 | Case 3 | Case 4 | Case 5 | Case 6 | Case 7 | Case 8 | Case 9 | Case 10 | Case 11 | Case 12 | Case 13 | Case 14 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Age | 3d 7h 12m | 42m | 2d 1h 47m | 1h 2m | 5d 20h 5m | 3h 1m | 1d 20h 48m | 1d 12h 46m | 1d 2h 45m | 20m | 18h | 5d 19m | 41m | 1h 20m |
| Maternal age (years) | 33 | 28 | 30 | 35 | 26 | 26 | 15 | 23 | 36 | 23 | 30 | 27 | 18 | 23 |
| Duration of PROM | 8d | 0 | 6d | 26h | 0 | 0 | 1d 2h | 8h | 0 | 0 | 0 | 1h | 0 | 0 |
| Delivery | C | C | C | ED | C | C | C | ED | C | ED | C | C | ED | DD |
| Gender | Male | Male | Male | Female | Male | Male | Female | Female | Female | Female | Male | Male | Female | Female |
| Apgar 1 and 5 minutes | 2-5 | 5-4 | 3-7 | 1-2 | 7-8 | 4-7 | 4-9 | 4-8 | 4-8 | 0-1 | 3-7 | 8-9 | 1-1 | 1-1 |
| Weight (g) | 590 | 1,050 | 740 | 710 | 2,000 | 910 | 1,840 | 750 | 850 | 520 | 1,290 | 2,350 | 890 | 1,480 |
| Height (cm) | 29 | 37 | 33 | 34 | 44 | 33 | 43 | 36 | 31.5 | 30 | 38 | 45 | 34 | 32 |
| Gestational age (weeks) | 25 | 29 | 27 | 26 | 41 | 26 | 32 | 25 | 27 | 25 | 27 | 32 | 25 | 31 |
| Maternal diagnostic | Hyp | PD | PROM Ch | PROM Ch | IGR | UTI Ch | PROM | PROM Ch | Severe Pre-eclam | PD UTI | Isoimm | Hydrops fetalis | PD | PD |
| Neonatal antibiotic treatment | Ampi Amika | - | Ampi Amika | - | - | - | Ampi Amika | Ampi Amika | - | - | Dicloxa Amika | - | - | - |
| Growth for gestational age | SGA | AGA | SGA | AGA | SGA | AGA | AGA | AGA | AGA | SGA | AGA | LGE | AGA | AGA |
AGA, appropriate for gestational age; Amika, amikacin; Ampi, ampicillin; C, Cesarean section; Ch, chorioamnionitis; d, days; DD, dystocic delivery; Dicloxa, dicloxacillin; ED, eutocic delivery; h, hours; Hyp, hypothyroidism; IGR, intrauterine growth retardation; Isoimm, isoimmunization; LGE, large for gestational age; m, minutes; PD, preterm delivery; Pre-eclam, pre-eclampsia; PROM, premature rupture of membranes; SGA, small for gestational age; UTI, urinary tract infection.
The causes of death in the remaining cases (cases 5 to 7 and 9 to 14) were not associated with infection. Four (29%) died due to causes related with their premature birth, three (21%) due to structural congenital defects not compatible with life, one due to hydrops fetalis of non-immunological origin with severe preeclampsia, and one due to maternal-fetal isoimmunization to Rh. All these cases had negative polymerase chain reaction results for C. trachomatis and Mycoplasma spp., with the exception of case 6, previously mentioned.
DiscussionSevere infections are still the main cause of neonatal morbidity and mortality in developing regions. Three-fourths of infant mortality cases occur during the first week of life, and in many cases, the cause of death remains unknown.23
Chlamydial infection in the perinatal and neonatal stage can cause many diseases. These include conjunctivitis, nasopharyngitis, pneumonitis, and less frequently, rhinitis, middle ear otitis, myocarditis, and encephalitis.4 However, the discovery of genetic residues of this intracellular bacterium in organic tissues is quite rare in the medical literature.24 In this study, chlamydial DNA was found in more than two organs in four cases (cases 1, 2, 3, and 4), which suggest the potential for a multivisceral infection by this pathogen. One notable instance was case 2 where the neonate was born prematurely by cesarean and died due to extensive pneumothorax. Case 2 did not have histopathological data for inflammation in the lungs or placenta; however, the repeated finding of chlamydial DNA in the brain and liver of this newborn presents a strong suspicion of systemic infection by C. trachomatis. Due to the lack of determination of histopathological criteria, this case suggests that the entire spectrum of C. trachomatis pathogenesis is still not completely known.
At no moment were diagnostic studies performed regarding Chlamydia or Mycoplasma infection in these patients, probably due to their short lifespan. Likewise, the mothers were also not screened for atypical pathogens during their pregnancy. The lack of C. trachomatis infection studies in these women was probably due to poor or no prenatal care: six (43%) women had no prenatal care, five (36%) had only two or three consultations, and three (21%) had five or six prenatal medical consultations.
The cause of death for case 8 was a systemic infection by Mycoplasma hominis. This pathogen has been shown to produce severe infection in fetuses and newborns, and can be vertically transmitted. Among the infections caused by Mycoplasma hominis, pneumonia, which evolves rapidly to bronchopulmonary dysplasia, and systemic infections with poor prognoses if not detected and treated in time25 stand out. This situation could also occur with the infections caused by C. trachomatis.
The polymerase chain reaction is the most sensitive and rapid method to detect microbial pathogens in clinical specimens, particularly, for Chlamydia and Mycoplasma, which are difficult to culture in vitro. The application of this method to clinical specimens has many potential pitfalls due to the presence of inhibitors and contamination. Further, the sensitivity and specificity of this assay is dependent on target genes, primer sequences, techniques used, DNA extraction procedures, and amplified product detection methods. However, the present investigation group had previously standardized the polymerase chain reaction applied in this study.24 Additionally, the positive samples in the assay were confirmed through real-time polymerase chain reaction, a method that offers many general technical advantages, including reduced probabilities of variability and contamination, online monitoring, and no requirement for post-reaction analyses.
The current capacity to detect infections such as C. trachomatis, Mycoplasma, and Ureaplasma (infections that impact the health of the most vulnerable populations: newborns and pregnant women), will allow for the identification of the high frequency by which these microorganisms produce membrane ruptures, premature births, and neonatal disease that can become severe and even fatal.6 Therefore, it is believed that, in the future, it will be necessary to reassess the public policies on prenatal care.
Five of 14 cases in this series received empirical antimicrobial treatment. The antibiotic scheme administered to two of the newborns that died with systemic infections by C. trachomatis (cases 1 and 3) consisted of an aminoglycoside and a beta-lactam, treatment that is usually used when there is suspicion of congenital infection, since it covers almost all etiological possibilities of neonatal infection acquired in utero.26 However, the Chlamydia and Mycoplasma genera are not included in their antimicrobial spectrum.27 Ocular prophylaxis can fail to prevent neonatal chlamydial conjunctivitis, and does not prevent colonization or infection in the lungs; the only means of preventing chlamydial infection in the newborn is treating the infected mother.
Case 1 did not have bacterial or fungal development in the performed cultures, and case 2 was classified as contaminated because the cerebrospinal fluid sample was obtained postmortem and developed the polymicrobial species Providencia rettgeri and Enterobacter gergoviae, which are not known as producers of neonatal infections. Case 3 was delivered by cesarean section with six days of premature membrane rupture and maternal chorioamnionitis. At birth, a sample of umbilical cord blood was obtained for culture, which showed the development of Staphylococcus haemolyticus. This bacterium is part of the cutaneous flora and is not a pathogen that produces congenital neonatal infections.28 Additionally, treatment with ampicillin and amikacin does not cover Staphylococcus haemolyticus, which is characteristically multiresistant. These premises support the conclusion that those samples were contaminated.
The serotypes of C. trachomatis with high invasive capacity can invade diverse tissues of adult individuals and cause lymphogranuloma (L1, L2, L2a, and L3). In this study, it was shown that the C. trachomatis infection in case 1 was due to genotype D, which is one of the highest prevalence serotypes worldwide in genitourinary infections of the sexually active population.27 Genotype D is one of the few C. trachomatis serotypes with a cytotoxin that has great homology to the family of large cytoxins (LCTs) produced by Clostridium difficile. These LCTs cause diverse cytopathic effects in their host cells because their glucosyltransferase activity modifies the intracellular regulatory molecules such as the GTP binding molecule of the Ras superfamily.29 Additionally, LCTs generally interfere with the organization and dynamics of actin in both the cytoskeleton and intracellular trafficking.30 The cytopathic effect produced by the cytotoxin of C. trachomatis serotype D results in the rounding of infected cells due to depolymerization of actin. This could help explain, in part, the capacity of serotype D to infect and disseminate to diverse organs in those newborns in whom chlamydial DNA was found. However, the mechanisms through which C. trachomatis infection is disseminated to different organs are yet to be identified.
This study presented the possibility of infection by C. trachomatis to various organs of fetuses and newborns, and that this might have been associated with infant mortality. However, further studies are needed to confirm this finding.
The finding of chlamydial DNA in more than one organ of autopsy may not be the most accepted way for diagnosing systemic infection or establishing cause of death. However, in the authors’ opinion, the discovery of three different C. trachomatis DNA sequences by end-point and real-time polymerase chain reaction and identification in one case of the C. trachomatis genotype involved provides sufficient evidence for further investigations using additional and improved resources. Immunohistochemistry would strengthen the evidence presented here, but unfortunately, immunohistochemical stains were not performed in the present samples.
Conflicts of interestThe authors declare to have no conflict of interests.
Please cite this article as: Hernandez-Trejo M, Herrera-Gonzalez NE, Escobedo-Guerra MR, de Haro-Cruz MJ, Moreno-Verduzco ER, Lopez-Hurtado M, et al. Reporting detection of Chlamydia trachomatis DNA in tissues of neonatal death cases. J Pediatr (Rio J). 2014;90:182–9.