To explore possible genes related to the development of persistent pulmonary hypertension of the newborn (PPHN).
MethodsThe authors identified 285 single nucleotide polymorphisms (SNPs) of 11 candidate genes (BMPR2, EPAS1, PDE3A, VEGFA, ENG, NOTCH3, SOD3, CPS1, ABCA3, ACVRL1, and SMAD9), using an Illumina Asian Screening Array-24 v1.0 BeadChip Array. The FastLmmC and R package was used for statistical analyses. The chi-square test and Cochrane-Armitage trend test were used to compare the allele and genotype frequencies between the groups and to test the genetic models, respectively.
ResultsA total of 45 PPHN infants and 294 control subjects were analyzed. The most common cause of PPHN was meconium aspiration syndrome. Among the 285 SNPs, 17 SNPs from 6 candidate genes (BMPR2, EPAS1, PDE3A, VEGFA, ENG, and NOTCH3) were significantly associated with PPHN (P < 0.05). After using the Bonferroni correction (P < 0.00018), only the rs17034984 SNP located in intron 1 of the EPAS1 gene remained significantly different between the PPHN and control subjects (P = 0.00014). The frequency of the TC/TT genotype of rs17034984 in the gene with the dominant model was significant in the patients with PPHN (OR = 5.38, 95% CI: 2.15–13.49). The T allele frequency of rs17034984 in the gene showed a significant difference compared with the control subjects (OR = 4.89, 95% CI: 2.03–11.82).
ConclusionsThe present study suggests that the rs17034984 variant of EPAS1 gene is associated with PPHN.
Persistent pulmonary hypertension of the newborn (PPHN) is the most serious complication of several neonatal respiratory diseases, leading to adverse outcomes such as asphyxia, chronic lung disease, neurodevelopmental sequelae, and death, despite the use of modern therapeutic modalities for treating PPHN such as high-frequency oscillatory ventilation, inhaled nitric oxide, and extracorporeal membrane oxygenation.1,2 Although the primary etiologies of PPHN are various factors such as chronic hypoxia, meconium aspiration syndrome, birth asphyxia, sepsis, infection, and respiratory distress syndrome,1,3,4 a genetic contribution to PPHN has been increasingly recognized. Previous studies have shown that SNPs in the CRHR1 (rs4458044), CPS1 (rs41272673, rs4399666, rs2287599, rs192759073, rs1047883, and rs2229589), EDN1 (rs2070699), and NOTCH3 (rs1044008) genes are susceptibility loci associated with PPHN.5–9 It has also recently been reported that mutations in a few genes such as TTLL3 and ITGAM10 were associated with PPHN. TBX4 variant has also been found in relation to neonatal and pediatric pulmonary hypertension.11
In this study, the authors aimed to determine any associations between SNPs of previous PPHN candidate genes and to perform a validation of any genetic associations found to elucidate a possible biological involvement of the protein(s) encoded from the gene(s) in the etiology of PPHN.
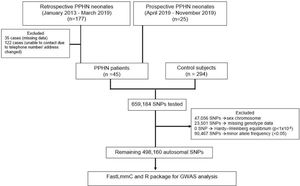
Materials and methodsStudy populationThe authors recruited 45 patients with PPHN who were hospitalized at Hat Yai hospital, one of the biggest hospitals in Southern Thailand where the prevalence of PPHN is relatively high, from 2.6 to 2.8 per 1000 live births, and constituting ∼30% of the all-cause mortality rate.1,12 Retrospective data of PPHN infants discharged from the hospital between January 2013 and March 2019, born to mothers with a gestational age of ≥ 35 weeks and birth weight of ≥ 2000 g, were retrieved from the hospital computer data system. The authors recruited 20 patients from the retrospective data. The authors also recruited additional PPHN neonates who were hospitalized at the hospital between April 2019 and November 2019 (Figure 1). The authors excluded patients with congenital structural cyanotic heart diseases and other congenital disorders such as congenital diaphragmatic hernia and birth defects due to chromosomal disorders. For a healthy control group, the authors recruited 59 neonates who were born at the hospital between April 2019 and November 2019 and 235 adults who participated in the Thai SNP database project of Ramathibodi Hospital, Bangkok, Thailand. A summary of subjects involving in the study is shown in Figure 1 and baseline characteristics are presented in supplementary Table 1. This study was approved by the Research Ethics Committee of the Hat Yai Hospital (EC No. 24/2562).
Study definitionsThe diagnostic criteria for PPHN were presentation shortly after birth with refractory hypoxemia plus one of the three following conditions: 1) documented pulmonary hypertension, as defined by echocardiographic evidence of elevated pulmonary pressure (right to left or bidirectional shunt) and/or 2) a pre-to-postductal partial pressure of oxygen gradient equal to or greater than 20 mm Hg, and/or 3) a pulse oximetry oxygen saturation (SpO2) gradient equal to or greater than 10%.13
Genomic DNA extractionGenomic DNA was extracted from a 200 μL sample of peripheral blood using a Qiagen DNA Mini kit (Qiagen, Hilden, Germany). The quantity and quality of the DNA samples were evaluated using an Eppendorf BioPhotometer plus (Eppendorf AG, Hamburg, Germany) and automated electrophoresis (Agilent 2200 TapeStation system, Santa Clara, California, United States).
Genotyping and quality controlA total of 659,184 single nucleotide polymorphism (SNPs) probes specific to several regions across the genome were fabricated on an Illumina Asian Screening Array-24 v1.0 BeadChip Array. SNP genotyping using an Illumina Infinium HTS Assay platform (Illumina, San Diego, California) was carried out and 612,128 (93%) SNPs were successfully genotyped. For quality control, the authors applied a low call rate < 98%, a Hardy–Weinberg equilibrium calculations p-value < 1 × 10−8, and minor allele frequency (MAF) of each SNP <0.05 to exclude data with low quality. After the quality control exclusion, 498,160 autosomal SNPs were left for the genome-wide association study (GWAS) analysis. The authors selected the SNPs in the genes related to PPHN and pulmonary arterial hypertension (PAH). Finally, 285 SNPs of 11 candidate genes (BMPR2, EPAS1, PDE3A, VEGFA, ENG, NOTCH3, SOD3, CPS1, ABCA3, ACVRL1, and SMAD9) were analyzed.
Immunohistochemistry (IHC) detection of HIF-2α in umbilicus and lung tissuesFor the validation study, the authors evaluated the expression of hypoxia-inducible factor 2-alpha (HIF-2α), a protein encoded from the EPAS1 gene, in the pulmonary vascular endothelium of lung tissues obtained from deceased PPHN patients (n = 3) compared to normal adult lung tissue (n = 3). In addition, the authors carried out IHC staining of HIF-2α in the umbilicuses of 8 healthy newborns to evaluate the expression of the protein in the umbilical artery and vein. Paraffin-embedded umbilical and lung tissues were stained with hematoxylin and eosin to examine histopathology of the tissues. Then, IHC using anti-HIF-2α antibody (ab199, Abcam, UK) with dilution 1:100 v/v was performed on VENTANA BenchMark ULTRA platform (Ventana Medical Systems Inc., USA) to examine for HIF-2α expression.
Statistical analysisThe authors used FastLmmC (v2.02.20121124) and the R package for GWAS (v. 3.3.3) for statistical analysis. Regional plots were generated using LocusZoom v.0.4.8. The allele and genotype frequencies of the candidate genes were calculated by the genotypes of the subjects. The chi-square test was used to compare the allele and genotype frequencies between the groups. When comparing the genotype frequency of each significant SNP between groups, the authors used the Cochrane-Armitage trend test between all genotypes. Odds ratio (OR) and 95% confidence intervals (CI) were calculated to indicate the degree of association between groups.
ResultsAssociation of SNPs in previously reported candidate genes and PPHN/ PAHThe authors analyzed 285 SNPs in 11 candidate genes, including BMPR2, EPAS1, PDE3A, VEGFA, ENG, NOTCH3, SOD3, CPS1, ABCA3, ACVRL1, and SMAD9, that were found to be associated with the pathogenesis of PPHN/PAH in previous studies. A p-value of < 0.05 was considered significant. The authors found that 17 SNPs in 6 candidate genes: BMPR2, EPAS1, PDE3A, VEGFA, ENG, and NOTCH3, were significantly associated with PPHN (Table 1). Of these, rs17034984 in the EPAS1 gene was the only one to remain significant after the Bonferroni correction (P < 0.00018).
Candidate genes related to the pathogenesis of PPHN (17 SNPs from 6 candidate genes).
The genotype and allele frequencies of candidate gene polymorphisms are summarized in Supplementary Table 2. Various models of inheritance (codominant, dominant, and recessive) were tested and genetic effects were calculated for each genotype compared to the reference genotype in each model. The authors found that the frequency of the TC/TT genotype of rs17034984 in the EPAS1 gene with the dominant model was significant in the patients with PPHN (OR = 5.38, 95% CI: 2.15–13.49). In addition, the allele frequency of the minor allele T of rs17034984 in the EPAS1 gene showed a significant difference compared with the control subjects (OR = 4.89, 95% CI: 2.03–11.82).
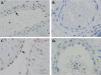
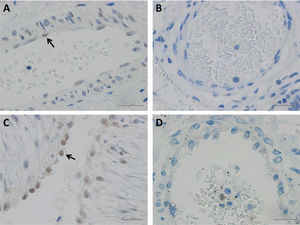
HIF-2α immunohistochemistry staining of the vascular endothelium of umbilicus and lung tissuesHIF-2α protein was expressed in lung tissues from 2 infants with PPHN. Localization of HIF-2α was seen in the nuclei of endothelial cells of the pulmonary arteries (Figure 2A). However, there was one infant with PPHN in whom HIF-2α protein expression was not detected (Figure 2B). The HIF-2α protein was also expressed in the endothelial cells of the umbilical arteries and veins from the 8 normal newborns (Figure 2C). The HIF-2α protein was not expressed in normal adult lung tissues and endothelial cells of the pulmonary vessels (Figure 2D). Determination of protein expression was scored by two pathologists (intensity of staining: 0, no staining; 1, weak; 2, moderate; and 3, strong). In addition, semiquantitative analysis of the IHC images was performed using ImageJ Fiji software (version 2.1.0/1.53C), following an integrated protocol.14 Expression of HIF-2α in different tissues were shown in Table 2.
A, Positive HIF-2α was detected in the nucleuses of few endothelial cells (arrow) of bronchial vessels in PPHN patients; B, Endothelial cells of bronchial arterioles stained negatively for HIF-2α; C, Positive HIF-2α was detected in the nucleuses of many endothelial cells (arrow) of umbilical cord vessels in healthy newborns; D, Immunostaining of HIF-2α stained negatively in adult bronchial vessels. Scale bar = 20 μm.
Expression of HIF-2α protein in different tissues.
Note: intensity of staining: 0, no staining; 1, weak; 2, moderate; and 3, strong.
PPHN is the most serious respiratory disorder in Thailand and has a high mortality rate. Its high prevalence in the study's institution led the authors to hypothesize whether genetic factors play an important role in the development of PPHN. Using SNP analysis, the authors found that rs17034984 in the EPAS1 gene showed a significant association with PPHN in the studied population. In addition, the authors found that the T allele of rs17034984 in the EPAS1 gene is a possible predisposing factor for PPHN and the TC/TT genotype with the dominant model could be a predictor of PPHN.
The rs17034984 SNP is located in intron 1 of the EPAS1 gene. The EPAS1 gene, located on the short arm of chromosome 2 (2p21), encodes endothelial PAS domain-containing protein 1, which is also known as hypoxia-inducible factor 2 alpha (HIF-2α) protein.15 This protein is a subunit of the HIF protein complex, which plays a crucial role in the body's ability to adapt to changing oxygen levels. HIF-2α is a transcription factor involved in the induction of oxygen-regulated genes. It regulates erythropoiesis and physiological responses to hypoxia through the HIFs signaling pathway.16 In hypoxic conditions, the HIFs protein is translocated to the nucleus to bind the hypoxic response element (HRE) of target gene promoters and subsequently initiate gene transcription. When oxygen levels rise, HIFs are hydroxylated by prolyl hydroxylase domain proteins (PHD), leading to interaction with other proteins, and with the addition to ubiquitin results in HIFs degradation.15 Previous studies reported that the EPAS1 gene polymorphisms were associated with adaptation to hypobaric hypoxia in the Tibetan population.17,18 In addition, HIF-2α regulates the vascular endothelial growth factor (VEGF) expression and seems to be implicated in the development of blood vessels and the tubular system of the lung. An animal model study also showed that the gain-of-function of HIF-2α was related to the development of pulmonary vascular remodeling.19 Another study found that chronic hypoxia could induce HIF-2α stability, leading to increased arginase expression and dysregulating normal vascular nitric oxide homeostasis.20 In addition, endothelial HIF-2α was a major mediator in the development of hypoxic-induced pulmonary hypertension, and its functions could induce pulmonary hypertension through an increased expression of endothelin-1, encoded from the EDN1 gene and a concomitant decrease in vasodilatory apelin receptor signaling.21 Gene network analysis showed that EPAS1 gene is co-expressed with EDN1 gene. The Gene network of 11 candidate genes in the present study study (BMPR2, EPAS1, PDE3A, VEGFA, ENG, NOTCH3, SOD3, CPS1, ABCA3, ACVRL1, and SMAD9) and other genes that had been shown to be associated with PPHN is shown in Supplementary Figure 1.
In the present study, 10 SNPs of the EPAS1 gene showed a significant difference between patients with PPHN and the controls (Table 1); however, rs17034984 was the only SNP that remained associated with PPHN after the Bonferroni correction. The T allele on rs17034984 was a risk factor for PPHN and the TC/TT genotype with the dominant model was significantly higher in patients with PPHN. However, to date, there have been few studies regarding HIF-2α in neonates. One study reported that HIF-2α was highly expressed in the lungs of neonates with a congenital diaphragmatic hernia and was possibly correlated with PPHN in neonates.22 Taking these findings together, the authors believe that a genetic variant of the EPAS1 gene is possibly associated with PPHN.
To demonstrate that expression of the HIF-2α protein (i.e., expression of the EPAS1 gene) in pulmonary vascular endothelial cells is involved in PPHN pathophysiology, the authors carried out IHC staining in pulmonary vascular endothelial cells and other tissues. The authors found that HIF-2α protein was expressed in the nucleus of pulmonary vascular endothelial cells of some PPHN patients, whereas there was no HIF-2α protein expression in normal adult lung tissues. Additionally, the authors observed HIF-2α protein expression in fetal umbilical vascular endothelial cells, possibly as a result of exposure to a hypoxic environment during physiological development. During in utero development, expression of both HIF-1α and HIF-2α are relatively high in the fetal lungs due to fetal exposure to a hypoxic environment.23–25 HIF-1α expression is predominant in respiratory epithelial cells, while HIF-2α is predominant in vascular endothelial cells and type II pneumocytes.26 Due to a limited number of PPHN lung samples, the authors could not perform statistical analysis to make a solid conclusion that HIF-2α was significantly higher expressed in neonatal PPHN lungs. However, the present study's results suggest that more studies are needed to further explicate the role(s) of HIF-2α in the pathophysiology of PPHN.
There are some limitations to the present study. First, the number of neonates with PPHN recruited into this study was relatively small. Second, the number of SNP probes fabricated onto the microarray might not cover all SNPs associated with the pathogenesis of PPHN. Third, although the present study carried out a preliminary validation study, using IHC staining to detect HIF-2α protein expression in lung tissues from neonates with PPHN, the authors could not compare the study's findings with normal lung tissues from neonates because autopsies of deceased neonates were not routinely performed in the study's center. Instead, the authors carried out IHC staining to detect HIF-2α protein expression in normal adult lung tissues. Concerning potential bias in adult control subject selection due to unknown medical history in their neonatal period, this concern was deemed minimal. Since the prevalence of PPHN is ∼3 per 1000 livebirth, which means the vast majority of individuals do not have PPHN.
In conclusion, in this study, the authors explored novel candidate SNPs that might be involved in the pathophysiology of PPHN. The authors’ analysis revealed that rs17034984 in the EPAS1 gene is a possible genetic susceptibility locus associated with PPHN. In combination with IHC staining to detect HIF-2α protein expression, the authors found that HIF-2α, encoded by the EPAS1 gene, might be a factor associated with PPHN via pulmonary vascular remodeling related to different oxygen levels in fetuses. However, further studies are needed to confirm the authors’ findings and to further explore the pathophysiological mechanism(s) underlying this association.
FundingGraduate School and Faculty of Medicine Foundation, Prince of Songkla University, and Medical Genetics Center, Division of Genomic Medicine and Innovation Support, Department of Medical Sciences, Ministry of Public Health, Thailand.
The authors thank Dr. Jakris Eu-ahsunthornwattana of the Department of Community Medicine, Faculty of Medicine Ramathibodi Hospital, Mahidol University for allowing the authors to use the Thai SNP database. The authors thank Mr. David Patterson of the Office of International Relations, Faculty of Medicine, Prince of Songkla University for editing the manuscript.