To describe the genotypic variability of group A rotavirus (RVA) found in immunosuppressed and non-immunosuppressed pediatric patients treated at the Hospital de Clínicas da Universidade Federal do Paraná (HC-UFPR), Curitiba, Paraná.
MethodsA cross-sectional study was conducted with 1,140 stool samples collected from April, 2001 to December, 2008 in outpatients and hospitalized patients with acute gastroenteritis referred to the hospital. RVA diagnosis was performed through the latex agglutination method and enzyme immunoassay. Reverse transcription followed by multiplex hemi-nested polymerase chain reaction (PCR) and nucleotide sequencing were used for genotype characterization. Genotype combinations, clinical, epidemiological, laboratory data, and presence of hospital-acquired infections were reported.
ResultsA total of 80 rotavirus-positive stool samples were analyzed. The most frequent associations between genotypes G and P were: G4 P[8] (38.9%), G1 P[8] (30.5%), G9 P[8] (13.9%), G2 P[4] (6.9%), and G3 P[8] (1.4%). G2 P[4] was the most prevalent genotype after the vaccine implementation in the years 2006 and 2008. A total of 62,5% of infected children were aged less than 12 months. Of these, 55.6% had severe dehydration and 26.7% needed intensive care. A frequency of 12.5% of nosocomial infections was found. No correlation was observed between genotype and severity of infection in the study patients.
ConclusionRVA infections can be associated with severe clinical manifestations, and the surveillance of genotypic variability of this virus is crucial to monitor the emergence of new strains and the impact of the immunization in these patients.
Descrever a variabilidade genotípica do rotavírus grupo A (RVA) encontrado em pacientes pediátricos imunocompetentes e imunocomprometidos tratados no Hospital de Clínicas/Universidade Federal do Paraná (HC/UFPR), Curitiba, Paraná.
MétodosFoi realizado um estudo transversal com 1.140 amostras de fezes coletadas, de abril de 2001 a dezembro de 2008, em pacientes ambulatoriais e pacientes hospitalizados com gastroenterite aguda encaminhados ao hospital. As técnicas usadas foram o método da aglutinação do látex e imunoensaio enzimático para diagnóstico de RVA. Foi realizada transcrição reversa, seguida por PCR multiplex semi-nested e sequência de nucleotídeos para caracterização do genótipo. Foram relatadas as combinações genotípicas, dados clínicos, epidemiológicos,laboratoriais e a presença de infecções hospitalares.
ResultadosFoi analisado um total de 80 amostras de fezes positivas para rotavírus. As associações mais frequentes entre os genótipos G e P foram: G4 P[8] (38,9%), G1 P[8] (30,5%), G9 P[8] (13,9%), G2 P[4] (6.9%) e G3 P[8] (1.4%). O genótipo prevalente foi G2 P[4] depois da implementação da vacina nos anos de 2006 e 2008. Verificou-se que um total de 62,5% das crianças infectadas tinham idade abaixo de 12 meses. Destas, 55,6% tinham desidratação grave, e 26,7% precisaram de cuidados intensivos. Encontrou-se uma frequência de 12,5% de infecções hospitalares. Não se observou correlação entre o genótipo e a gravidade da infecção nos pacientes estudados.
ConclusãoAs infecções por RVA podem associar-se a manifestações clínicas graves e é crucial a vigilância da variabilidade genotípica desse vírus para monitorizar a emergência de novas cepas e o impacto da imunização nesses pacientes.
Group A rotaviruses (RVA) are the major etiologic agents of acute watery diarrhea in children aged less than 5 years worldwide. On a global scale, they are responsible for approximately 611,000 deaths per year, mostly in low-income countries.1
RVA infections remain an important cause of pediatric hospitalization, particularly in developing countries, where demographic and socio-economic factors are associated with increased mortality rates. Vaccination has a significant impact on the frequency of disease; nevertheless, severe infections persist, and the possible emergence of new genotypes must be considered. The diversity of rotavirus strains underscores the need for intensive strain surveillance; thus, the implementation of laboratory surveillance is critical to prevent outbreaks.2
Rotaviruses are classified into seven major groups (A through G), but most of infections are associated to rotavirus A, although groups B and C have been found in human illness. Among RVA, distinct genotypes (G and P outer capsid antigen) have been described, with G1P[8], G2P[4], G3P[8], G4P[8], and G9P[8] the most commonly identified worldwide and in Brazil.3
Several reports have demonstrated the importance of this pathogen as responsible for hospitalization of children with acute gastroenteritis (AGE). In Brazil, epidemiological findings suggest detection rates ranging from 12% to 42%.4 The Hospital de Clínicas da Universidade Federal do Paraná (HC-UFPR) is a tertiary center that receives patients referred from Curitiba and the metropolitan region. Analyzes of the cause of all cases of gastroenteritis admitted to the HC-UFPR have shown that RVA is the most frequently found pathogen (20%) in the studied population.5
In 2006, two rotavirus vaccines became available, a monovalent rotavirus vaccine (Rotarix®, GlaxoSmithKline Biologicals Inc) and a pentavalent rotavirus vaccine (RV5; RotaTeq®, Merck & Co., Inc.). Both vaccines are recommended by the World Health Organization (WHO) and have been used in several countries, and they have demonstrated a significant reduction of hospitalization and mortality due to rotavirus gastroenteritis.6,7
Brazil was one of the first countries to introduce universal vaccination against RVA, Rotarix®, which has been provided free through the public health system since March of 2006. The vaccine coverage in all the country in 2006 and 2007 was 60% and 75%, respectively.8 However, the South and Southeast Regions had the highest vaccine coverage, and it the largest reduction in the rate of hospitalization of children due to AGE was observed there.9
Previous analysis conducted in 2009 at this hospital to assess the impact of vaccination against RVA showed a reduction of 54.2% and 39.4% in medical consultations for children less than 12 months old and between 12 and 60 months, respectively. Furthermore, there was a reduction of 43.9% and 33.3% in the number of hospitalizations for gastroenteritis in children under 12 months and aged 12–60 months, respectively, considering the coverage of around 80% in the abovementioned period (unpublished data). Epidemiological surveillance for RVA diarrheal illness was established in the country to monitor the genotypic diversity of circulating RVA, as well as the rise of emerging and re-emerging RVA strains .10
Several studies involving the genetic variability of RVA have been published in Brazil; nonetheless, the majority of these were conducted in the Central and Southeast Regions, and the information about other regions is scarce. This study aimed to describe RVA genotypic variability over an eight-year period, and to assess clinical and epidemiological features of infected patients, as well as the impact of the immunization program on viral genetic diversification.
MethodsSamples and study designThis was a cross-sectional study that involved the examination of 1,140 stool samples of outpatients and inpatients with acute gastroenteritis referred to HC–UFPR. The patients were admitted to pediatric wards or to the hematopoietic stem cells transplantation (HSCT) unit. The stool samples were collected from April of 2001 to December of 2008, and were sent to the virology laboratory for RVA detection and posterior genotyping studies. Medical records of infected patients were reviewed, and the clinical data were collected using specific forms.
This study was approved by the Ethics of Research on Human Beings Committee of the HC-UFPR, under registration No. 4441.023/2002-04.
Criteria for the classification of severity of dehydrationDehydration was classified as mild, moderate, or severe, and evaluated on a clinical dehydration scale for children as previously reported.11
Detection of viral antigenFecal samples were initially tested for group A rotavirus antigen by screening tests - LA, (Virotect Rota kit–Omega Diagnostics or Rotascreen kit–Microgen Bioproducts) and EIA (EIARA kit–Biomanguinhos or Rotascreen II kit–Microgen Bioproducts), according to the manufacturer's instructions. The performance of these methods was analyzed and their results were compared to those previously reported.12 Positive samples were sequentially analyzed by molecular methods.
Viral genomic RNA extraction and multiplex hemi-nested RT- PCRGenomic RNA was extracted using aliquots of 200μL of fecal suspension (10% wt/vol) and silica filter, in accordance with the process previously described.13 The RNA obtained was analyzed by a multiplex hemi-nested real time polymerase chain reaction (RT-PCR) to define the viral genotype, using previously described methods.13 Briefly, the RNA was reverse transcribed and amplified by using specific primers corresponding to a conserved nucleotide sequence of the VP4 and VP7 genes, fragments of 876bp and 904bp, respectively.13 The amplified fragment was used as a template to a second PCR, using a combined typing scheme of the pool of primers to identify VP7: pool A (G1, G2, G3, G4, and G5), pool B (G8, G9, and G10), and pool C (G6 and G11); and to identify VP4: pool A [P4], [P6], [P8], [P9], and pNCDV [P1] genotypes. The results obtained were confirmed with individual primers for the identified genotype.
Sequencing, assembling and nucleotide sequence comparisonSamples that were positive in the first-step PCR, and which could not be genotyped by multiplex nested PCR were analyzed by nucleotide sequencing. The PCR products were purified using Invisorb® Spin PCRapid kit (Invitek Inc–USA), after both DNA strands were directly sequenced as described in the Thermo Sequenase kit (USB Inc – Ohio, USA) manual. BigDye® Terminator method was used on an ABI 3100 (Applied Biosystems Inc – USA). Specific primers from the first and second PCR were used to detect the RVA.
The BioEdit Sequence Alignment Editor was used to assemble the fragments into the most likely sequence.14 A set of VP4 and VP7 segment sequence was retrieved from Genbank,15 comprising representative RVA genotypes. Nucleotide differences were quantified using the MegaAlignTM software (DNASTAR®, Inc - USA).
All molecular analysis and sequencing reactions were performed at the virology laboratory of HC/UFPR.
Statistical analysesStatistical analysis was performed using the chi-squared test or Fisher's exact test, as appropriate. The tests were performed using GraphPad Prism version 5.0 for Windows (GraphPad Software – San Diego, California, USA). Only two-tailed tests were used. A p-value of < 0.05 was considered statistically significant.
ResultsDuring the study period, 179 (179/1,140 - 15.7%) samples were positive for RVA; of these, 80 (80/179 – 44.7%) had enough samples for further analysis, and were selected for the performance of multiplex hemi-nested RT-PCR for genotypes determination and nucleotide sequence, when necessary. 72 samples (72/80 - 90%) were RT-PCR positive for RVA; of these, 78% (56/72) were from hospitalized patients.
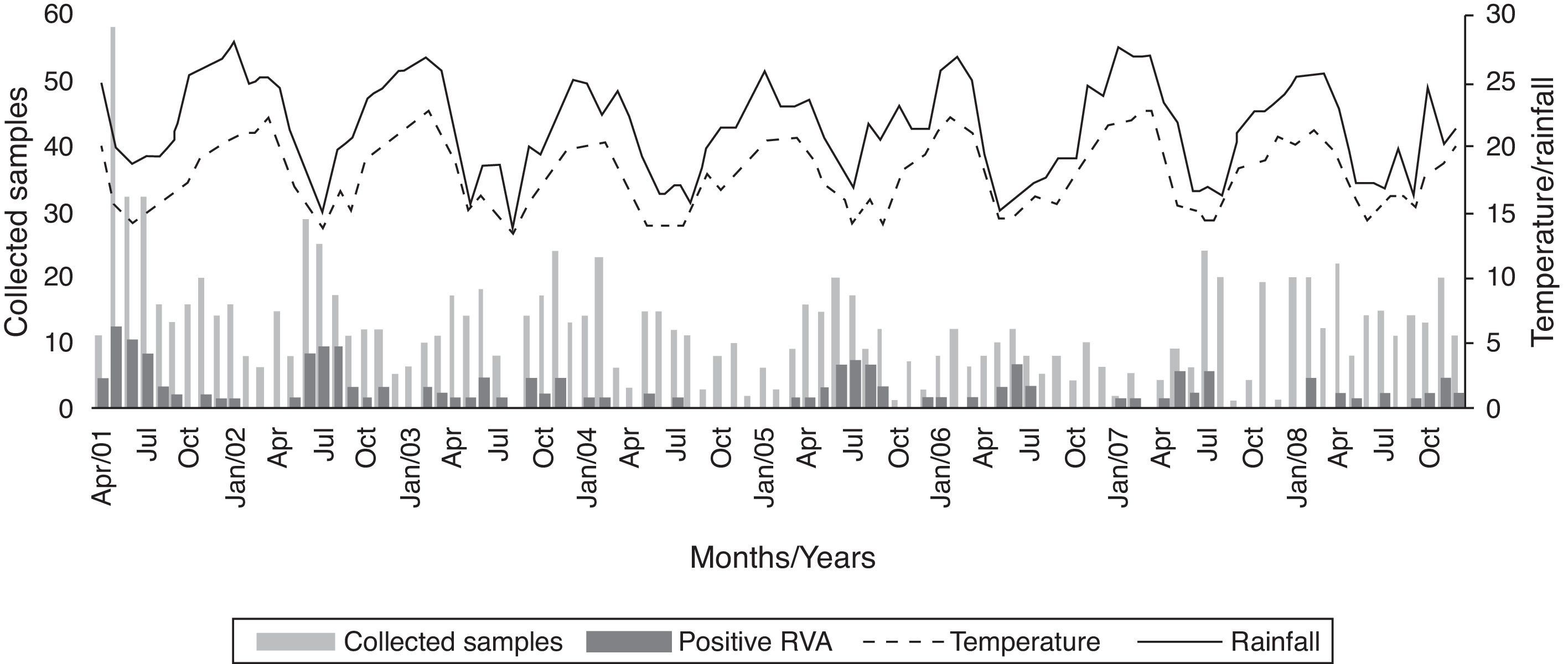
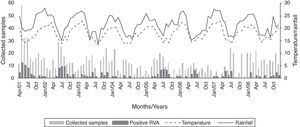
Fig. 1 shows the distribution of RVA during the eight-year study and its relation to monthly average temperature (°C) and rainfall (mm).
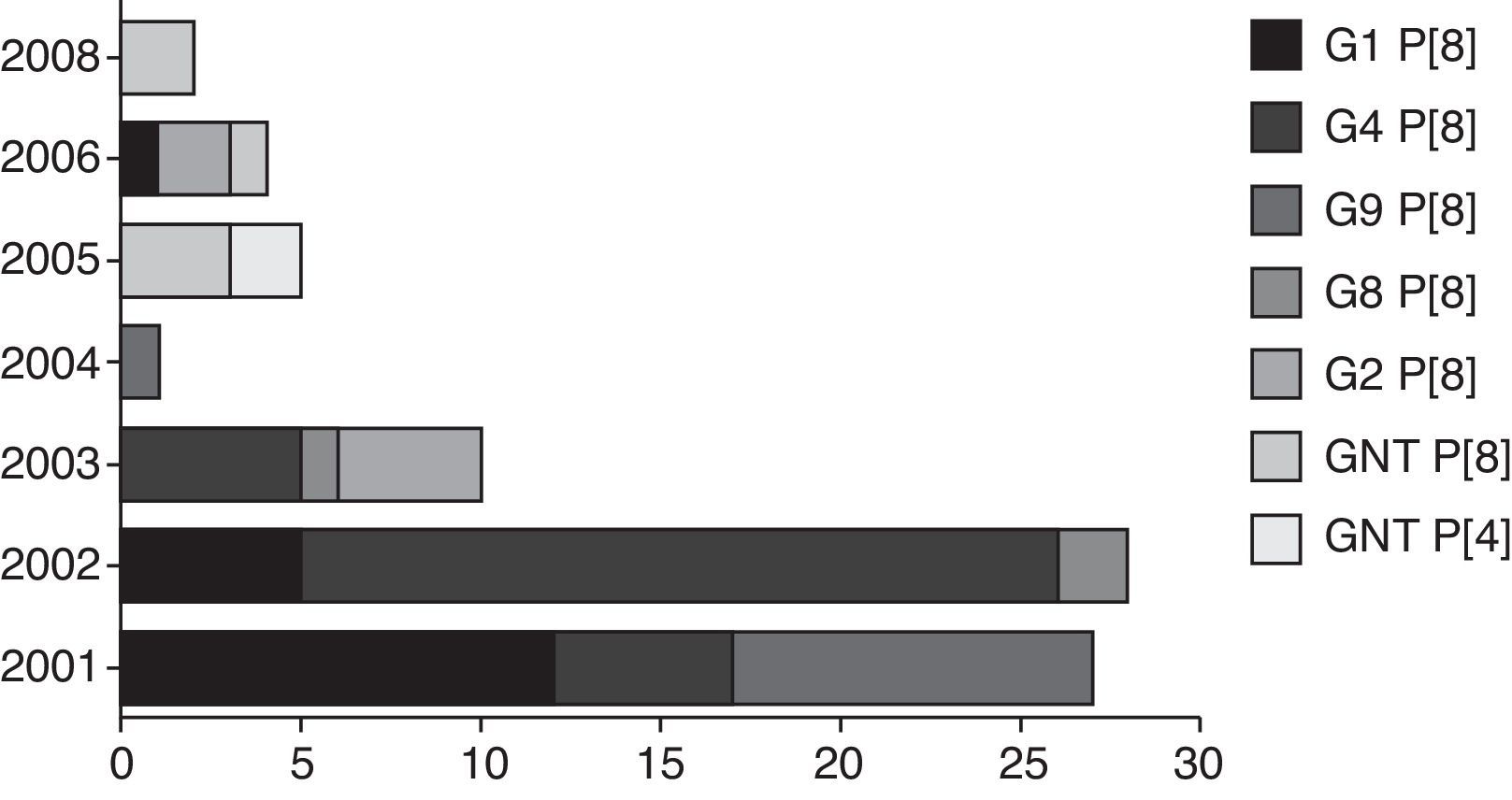
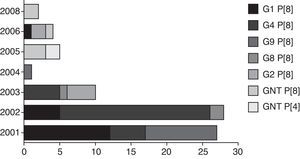
Sixty-six (66/72 – 91.6%) samples were genotyped. The genotypes found were G4 P [8] (28/72 – 38.9%), G1 P [8] (22/72 – 30.5%), G9 P [8] (10/72 – 13.9%), G2 P [4] (5/72 – 6.9%), and G3 P[8] (1/72 – 1.4%). Six samples could not be sequenced, probably because the primers used did not correspond to genotype investigated; also, these samples presented a weak band in the agarose gel, undermining the quality of sequencing reactions performed. Differences in G and P genotype distribution were detected in distinct years, reflecting the yearly change in epidemiology of human rotavirus, with an alternation between genotypes every one or two years (fig. 2). No mixed RVA infections were detected. After the implementation of the vaccination program, only G2 P [4] and GNT P [8] genotypes were found.
Demographic and clinical dataA total of 69 (69/80 - 86.2%) medical records were reviewed. 65% of the patients were male. The median age of patients was nine months (IQR, 6 - 16.5 months), and most cases occurred in patients aged < 12 months. Despite the broad frequency of patients with underlying diseases, a total of 51% (37/72) of the patients were admitted primarily due to severity of diarrhea. Regarding the vaccination status of patients admitted after 2006, only two patients reported previous RVA immunization: one received the complete scheme and the other only the first dose; both were non-immunosuppressed patients.
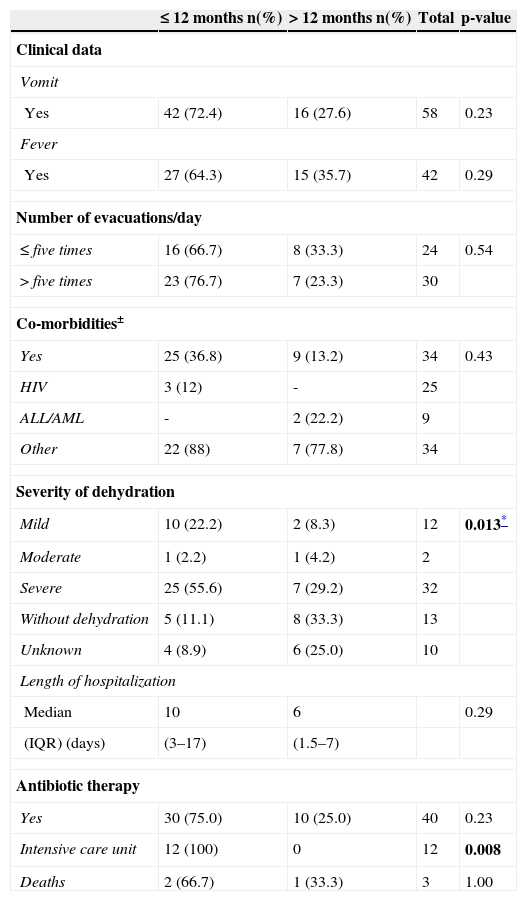
For clinical and laboratory data analysis, the patients were divided into two groups according to age: children ≤ 12 months (65%, 45/69) and children > 12 months (35%, 24/69). Comparison of the clinical ward showed a greater frequency of patients < 12 months in the intensive care unit (ICU) (p=0.008). A total of 64% of children presented dehydration, of which 78.3% were ≤ 12 months (p=0.01) (Table 1). Three (3.7%) children evolved to death, one related to gastroenteritis. No statistically significant differences were observed in the comparison between genotype and presence of dehydration (p=0.86).
Clinical and laboratory data of patients (n=69 patients) with acute gastroenteritis due to group A rotavirus, Hospital de Clínicas/Universidade Federal do Paraná, 2001 to 2008.
| ≤ 12 months n(%) | > 12 months n(%) | Total | p-value | |
|---|---|---|---|---|
| Clinical data | ||||
| Vomit | ||||
| Yes | 42 (72.4) | 16 (27.6) | 58 | 0.23 |
| Fever | ||||
| Yes | 27 (64.3) | 15 (35.7) | 42 | 0.29 |
| Number of evacuations/day | ||||
| ≤ five times | 16 (66.7) | 8 (33.3) | 24 | 0.54 |
| > five times | 23 (76.7) | 7 (23.3) | 30 | |
| Co-morbidities± | ||||
| Yes | 25 (36.8) | 9 (13.2) | 34 | 0.43 |
| HIV | 3 (12) | - | 25 | |
| ALL/AML | - | 2 (22.2) | 9 | |
| Other | 22 (88) | 7 (77.8) | 34 | |
| Severity of dehydration | ||||
| Mild | 10 (22.2) | 2 (8.3) | 12 | 0.013* |
| Moderate | 1 (2.2) | 1 (4.2) | 2 | |
| Severe | 25 (55.6) | 7 (29.2) | 32 | |
| Without dehydration | 5 (11.1) | 8 (33.3) | 13 | |
| Unknown | 4 (8.9) | 6 (25.0) | 10 | |
| Length of hospitalization | ||||
| Median | 10 | 6 | 0.29 | |
| (IQR) (days) | (3–17) | (1.5–7) | ||
| Antibiotic therapy | ||||
| Yes | 30 (75.0) | 10 (25.0) | 40 | 0.23 |
| Intensive care unit | 12 (100) | 0 | 12 | 0.008 |
| Deaths | 2 (66.7) | 1 (33.3) | 3 | 1.00 |
Data are shown as number (percentage), unless otherwise indicated.
ALL, acute lymphocytic leukemia; AML, acute myeloid leukemia; HIV, human immunodeficiency virus; IQR, interquartile range; n, number of samples.
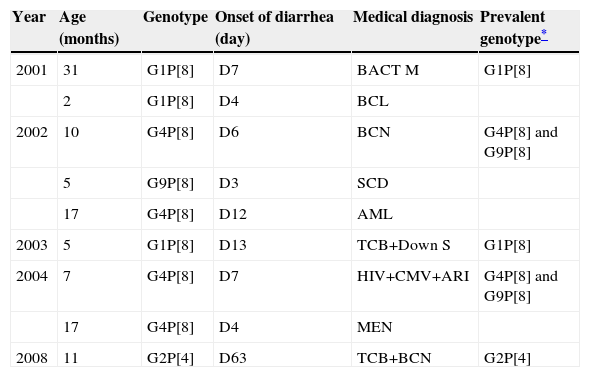
Nosocomial infections were considered those occurring 72hours after admission. From a total of 72 RVA-positive patients, 9 (12.5%) had diarrhea after this period. Of these, four patients were hospitalized in the pediatric emergency and five in pediatric infectious disease settings. The median length of hospitalization was seven days (IQR four to 12.5 days) (Table 2).
Group A rotavirus and hospital-acquired infections, 2001–2008.
| Year | Age (months) | Genotype | Onset of diarrhea (day) | Medical diagnosis | Prevalent genotype* |
|---|---|---|---|---|---|
| 2001 | 31 | G1P[8] | D7 | BACT M | G1P[8] |
| 2 | G1P[8] | D4 | BCL | ||
| 2002 | 10 | G4P[8] | D6 | BCN | G4P[8] and G9P[8] |
| 5 | G9P[8] | D3 | SCD | ||
| 17 | G4P[8] | D12 | AML | ||
| 2003 | 5 | G1P[8] | D13 | TCB+Down S | G1P[8] |
| 2004 | 7 | G4P[8] | D7 | HIV+CMV+ARI | G4P[8] and G9P[8] |
| 17 | G4P[8] | D4 | MEN | ||
| 2008 | 11 | G2P[4] | D63 | TCB+BCN | G2P[4] |
AML, acute myeloid leukemia; ARI, acute respiratory infection; BACT M, bacterial meningitis; BCL, bronchiolitis; BCN, bronchopneumonia; CMV, cytomegalovirus; D, day of hospitalization; Down S, Down syndrome; MEN, meningoccemia; SCD, sickle cell disease; TCB, tracheobronchitis.
The introduction of the RVA vaccine in the national immunization schedule contributed to a significant reduction in the frequency of this infection in the pediatric population. However, this pathogen can be associated with severe disease, and the surveillance of its genotypic variability is crucial to monitor the emergence of new strains circulating in humans.
The variation of G and P genotypes observed in different years highlights the mechanisms used by the RVA to escape immune selective pressure and thus maintain the pathogen in nature. Analysis of combinations of genotypes G and P type have demonstrated that genotype G1 P[8] was predominant in 2001 and 2003, similar to previous findings.13 The genotype G3 P[8] was identified in only one sample in 2003. Regarding genotype G4 P[8], its occurrence was detected in 2001, as the second most frequent genotype, and in 2002 it prevailed; similar results were reported in Paraguay.16 The genotype G9 P[8] was identified in this study in the years 2002, 2004, and 2005. According to some reports,17 this genotype is now circulating more widely. Genotype G2 P[4] has re-emerged since 2006, and it has become predominant in the samples analyzed in this study, as well as in other studies in Brazil and in other countries.18 In year 2007, no RVA was detected in the studied samples; however, some reports in Brazil showed the continuity of the detection of G2 P[4] genotype until 2008.19
These results demonstrate the ability of RVA genetic variation, and point out the need for constant surveillance of the circulating genotypes.20 After the introduction of universal vaccination in Brazil, the emergence of genotypes G2 P[4] and G9 P[8] was observed. The impact of these infections must be monitored in order to assess the effectiveness of the currently available vaccines. The predominance of genotype G2 P(4) in other countries using the Rotarix vaccine has been reported. It remains unclear whether this is due to selective pressure or changing epidemiology.21 However, an increase of the frequency of G2 P[4] genotype could be observed in periods previous to vaccine implementation and also in countries without RVA immunization,18,22 and thus its circulation was probably associated to the natural reemergence of this genotype.3,23
Similar to previous findings, in this study a significant decrease in the frequency of RVA infections in the pediatric population was observed since the implementation of immunization, and the positive cases observed concern patients with incomplete immunization status or immunosuppressed patients, who may not develop a complete immune response. After 2006, no RVA-positive samples were detected in outpatients.
RVA infections are most common in the wintertime in temperate regions, and year-round in tropical areas.24 In the present study, an increase in positive cases was observed in certain years, particularly during the colder months, in agreement with other findings.13 However, it has been found that the frequency of the disease varied throughout the year, suggesting that factors other than weather can influence the seasonality of this pathogen.25 Furthermore, in 2008, it was observed that RVA activity was spread throughout the entire year, peaking in the spring, which represented a delay of almost five months when compared to pre-immunization period. This was probably a result of a less susceptible population and, consequently, the virus required more time to spread.23
RVA infections were predominant in children aged 0–12 months according previous reports,26 and the clinical manifestations varied in intensity according to age and host immunity. The classical clinical picture of RVA infections is reported as the abrupt onset of vomiting, fever, followed by diarrhea, and leading to dehydration.25,27 It is worth noting that seven of the 12 patients affected by RVA infection who required hospitalization were admitted to the ICU with severe dehydration, did not have underlying diseases, and were younger than six months old.
A total of 49.2% of the hospitalized children was found to have moderate or severe dehydration, which corroborates the severity of this infection. However, no association between disease severity and genotype was found, demonstrating that other factors (mainly previous clinical conditions) may be associated with the severity and intensity of infections caused by RVA.28
It is worth mentioning the importance of RVA associated to hospital-acquired infections among children. Several factors, such as age, immune status, underlying disease, diagnostic and therapeutic interventions, season of the year, and duration of hospitalization may influence the acquisition of these infections. In addition to morbidity, these infections cause a major economic impact on developed and developing countries.29 The incidence of nosocomial infections in this study was 12.5%; other reports found rates ranging from 8% to 33%.30 All patients had serious underlying diseases and this infection may have contributed to the increase in severity. The genotypes found in these patients reflected the same genotype circulating in the community, highlighting the importance of measures for hospital infection control to prevent the spread of the pathogen in this environment.31
Epidemiological studies have demonstrated a clear correlation between RVA circulation period and increase in pediatric patient admissions. Although no specific antiviral therapy for RVA is available, the identification of infected patients is important to prevent nosocomial transmission, to understand the clinical impact of these infections, to guide therapeutic measures to prevent the inappropriate use of antibiotics, and to indicate the need for specific immunization.
In conclusion, this study has demonstrated the genetic variability of RVA during an extensive period of monitoring and the severity of these infections in pediatric patients. It has also emphasized the importance of ongoing laboratory surveillance to detect the emergence of new genotypes and to determine whether this is a consequence of the global program of immunization, and to assess its impact on pediatric health.
FundingFundação Araucária/State of Paraná, Brazil.
Conflicts of interestThe authors declare no conflicts of interest.
Please cite this article as: Pereira LA, Ferreira CE, Turchetto GD, Nogueira MB, Vidal LR, Cruz CR, et al. Molecular characterization of rotavirus genotypes in immunosuppressed and non-immunosuppressed pediatric patients. J Pediatr (Rio J). 2013;89:278–85.