to evaluate the accuracy of an antibody point-of-care lateral flow immunoassay (LFI - Wondfo Biotech Co., Guangzhou, China) in a pediatric population.
Methodschildren and adolescents (2 months to 18 years) with signs and symptoms suggestive of acute SARS-CoV-2 infection were prospectively investigated with nasopharyngeal RT-PCR and LFI at the emergency room. RT-PCR was performed at baseline, and LFI at the same time or scheduled for those with less than 7 days of the clinical picture. Overall accuracy, sensitivity and specificity were assessed, as well as according to the onset of symptoms (7-13 or ≥14 days) at the time of the LFI test.
ResultsIn 175 children included, RT-PCR and LFI were positive in 51 (29.14%) and 36 (20.57%), respectively. The overall sensitivity, specificity, positive and negative predictive value was 70.6% (95%CI 56.2-82.5), 96.8% (95%CI 91.9-99.1), 90.0% (95%CI 77.2-96.0), and 88.9% (95%CI 83.9-92.5), respectively. At 7-13 and ≥14 days after the onset of symptoms, sensitivity was 60.0% (95%CI 26.2-87.8) and 73.2% (95%CI 57.1-85.8) and specificity was 97.9% (95%CI 88.7-99.9) and 96.1% (95%CI 89.0-99.2), respectively.
ConclusionDespite its high specificity, in the present study the sensitivity of LFI in children was lower (around 70%) than most reports in adults. Although a positive result is informative, a negative LFI test cannot rule out COVID-19 in children.
The COVID-19 pandemic is the largest world health crisis of the century.1,2 The limitations of the effectiveness of the containment measures and the great impact in several aspects of society forced world leaders to make difficult decisions.3–5 The strategies to mitigate the pandemic include social distancing, use of face masks, hand hygiene, vaccines and the widespread use of accurate testing.6,7
Despite the large number of studies published since the beginning of the pandemic, the knowledge about SARS-CoV-2 infection tests accuracy is still evolving. As critical cases of COVID-19 were far more common in older adults and patients with comorbidities, most studies assessing the accuracy of diagnostic tests were performed in this population.8–11
Although children and adolescents with COVID-19 usually have a mild disease, severe cases occur,12 as is the case of the rare COVID-19-related multisystem inflammatory syndrome (MIS-C). This syndrome resembles Kawasaki-disease and can lead to multiorgan failure and even death.13–15 Also, children may play a role in SARS-CoV-2 spread, although the viral dynamics in the pediatric population are not fully understood.16–19 All this novel information justifies more precise definitions of the best accuracy of viral diagnostic tools, as an important public health preventive measure among the pediatric population.
The aim of this study was to assess the accuracy of an antibody LFI test in children and adolescents compared to RT-PCR in the acute care setting. The authors have also compared the test accuracy between infants and older children.
Materials and methodsThis is a prospective multicenter observational study with data collected in two hospitals, from May to November 2020. Pediatric patients (>2 months and <18 years) admitted at emergency rooms (ERs) or visiting outpatient clinics with signs or symptoms suggestive of COVID-19 (cough, fever, or sore throat) within 14 days of onset of symptoms were eligible. The exclusion criteria was the failure to collect samples for SARS-CoV-2 RT-PCR. Legal caregivers provided signed consent and authorization for their child's participation. The study was carried out according to the Declaration of Helsinki and Good Clinical Practice Guidelines, after approval by the study's Institutional Review Board (IRB No. 30749720.4.1001.5330).
The identification of SARS-CoV-2 through RT-PCR assays was performed in parallel for all participants. Nasopharyngeal and oropharyngeal swab sample collections were allocated in the same transport media with saline solution and RNAlater®, and RNA Stabilization Solution (Catalog number AM7021). RNA extraction was performed using MagMax™ Viral/Pathogenic Nucleic Acid Isolation (Applied Biosystems) in KingFisher Duo Prime System platform, (ThermoFisher, USA). The total reaction volume was 10 µL, with 5 µL of PathTM 1-Step RT-qPCR Master Mix, CG (Catalog Numbers A15299, AppliedBiosystems), TaqManTM 2019-nCoV Assay Kit v1 (Catalog Number A47532), and 5 µL of RNA. Reaction control had 5 µL (200 copies/µL) of TaqManTM 2019-nCoV Control Kit v1 (Catalog Number A47533). RT-PCR assay was performed on QuantStudio 5 (ThermoFisher Scientific, USA) and results were analyzed with QuantStudio™ Design & Analysis Software v1.5.1.
The SARS-CoV-2 Antibody Test (Wondfo Biotech Co., Guangzhou, China), was used to assess the presence of antibodies in two drops of blood from a finger-prick sample. This lateral flow assay detects immunoglobulins (Ig) G and M isotypes specific to the spike protein of SARS-CoV-2 receptor binding domain. The LFI assay reagent consists of colloidal gold particles coated with SARS-CoV-2 antigen-dye conjugate of receptor-binding domain (RBD; personal communication from the manufacturer). A positive result is characterized by the presence of human IgM and/or IgG SARS-CoV-2 specific antibodies in the blood sample. SARS-CoV-2 antibodies bind to antigen-dye conjugate and to colloidal gold complexes, defining a line test (T) in the kit's window, presenting as a dark-colored band. Instead, samples without SARS-CoV-2 specific antibodies will not display this dark-colored band, indicating a negative result. All valid tests presented a positive control band (C). If this control band was not visible, the test was considered invalid and the participant retested. If there was doubt about the result another researcher evaluated the result or the test was repeated.
RT-PCR sample collection was performed at baseline and LFI at the same time or scheduled for later in those with less than 7 days of clinical onset. All tests were performed through a standardized protocol by the trained study team. The laboratory team was blind to the LFI test results until the final analysis. However, when LFI was not performed at the inclusion, the research team had access to the RT-PCR test results.
Mean and standard deviation or median and interquartile range (IQR) were calculated for continuous variables and percentages for the categorical variables to describe the characteristics of the subject. Diagnostic accuracy was calculated using the epiR package (version 1.0-15). Sensitivity was characterized as the number of positive LFI test results, divided by the number of positive RT-PCR. Specificity was calculated as the number of negative LFI results divided by the total number of negative RT-PCR. Positive predictive value (PPV) was characterized as the number of positive LFI and RT-PCR test results (true positives) divided by the number of total LFI positives. Negative predictive value (NPV) was characterized as the number of negative LFI and RT-PCR test results (true negatives) divided by the number of total LFI negatives. The 95% confidence intervals for all estimates were calculated using the exact method.
To explore the accuracy according to the onset of symptoms, the authors calculated days of symptoms onset at the time of LFI testing, and assessed sensitivity and specificity at 7-13 days, and ≥14 days. The authors performed a sensitivity sub-analysis to compare the accuracy of LFI between infants (<2 years old) and older children (≥2 years old). Patients without both LFI and RT-PCR results were excluded from the analysis. All analyses were performed in R version 3.6.3
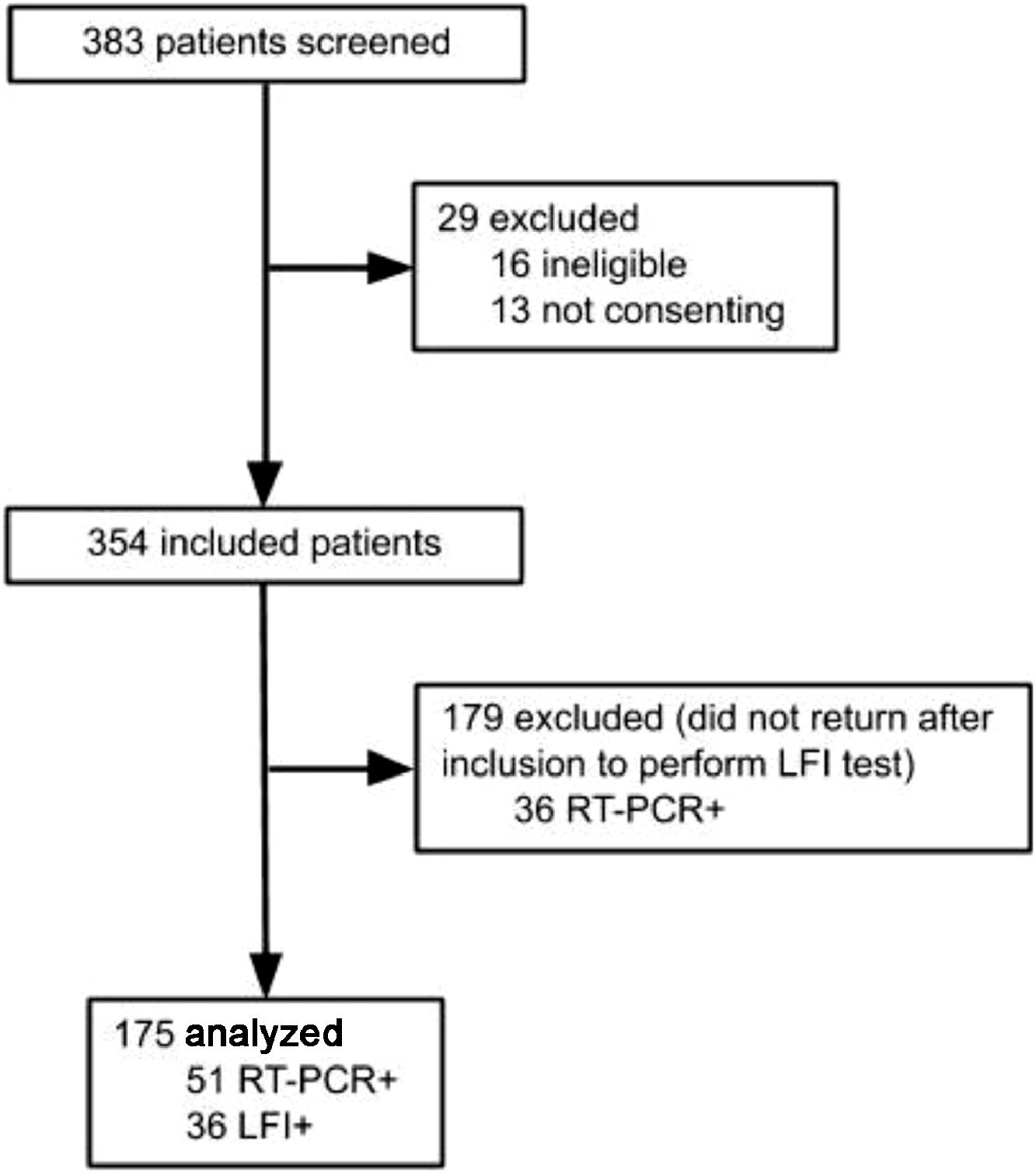
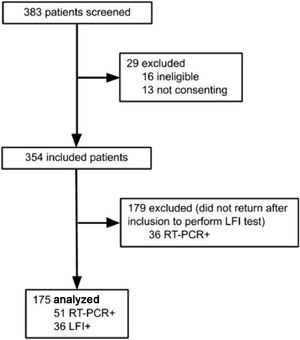
Results383 subjects were screened for the study, and 208 excluded: 16 for not meeting inclusion criteria, 13 for not consenting, and 179 did not perform LFI, as shown in Figure 1. A total of 175 participants were included in the final analysis, and SARS-CoV-2 infection was confirmed by RT-PCR in 51 (29.14%), while 36 (20.57%) were positive on the LFI. The authors observed 4 (2.28%) cases of negative RT-PCR with positive LFI. The comparison between included patients and those who did not perform LFI is shown in the Supplementary table 1.
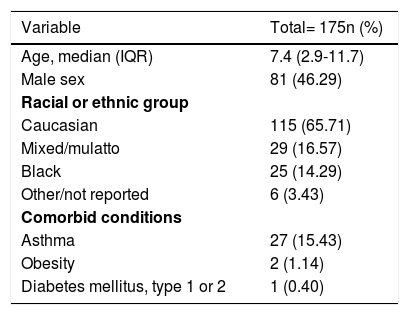
The most prevalent symptoms reported were cough (77.71%), coryza (72.57%), fever (66.29%), headache (59.43%), and stuffy nose (57.71%). Demographic and clinic details are presented in Table 1. The median (IQR) time from the onset of symptoms to RT-PCR assay was 3 (2-7) days. Twenty-eight patients performed RT-PCR after 7 days of onset of symptoms (23 with negative results). The median (IQR) time from the onset of symptoms to LFI testing was 16 (10-18) days.
Demographic and clinical characteristics of included subjects.
IQR, interquartile range.
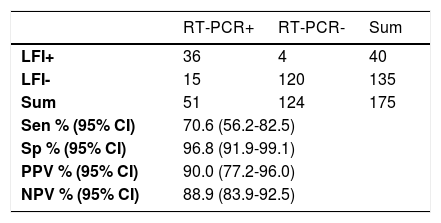
Overall sensitivity and specificity (≥7 days of symptom onset) was 70.6% (95%CI 56.2-82.5) and 96.8% (95%CI 91.9-99.1), respectively (Table 2). Positive predictive value was 90% (95% CI 77.0-96.0) and negative predictive value was 88.9% (95% CI 83.93-92.46) (Table 2).
Overall accuracy of the LFI test.
| RT-PCR+ | RT-PCR- | Sum | |
|---|---|---|---|
| LFI+ | 36 | 4 | 40 |
| LFI- | 15 | 120 | 135 |
| Sum | 51 | 124 | 175 |
| Sen % (95% CI) | 70.6 (56.2-82.5) | ||
| Sp % (95% CI) | 96.8 (91.9-99.1) | ||
| PPV % (95% CI) | 90.0 (77.2-96.0) | ||
| NPV % (95% CI) | 88.9 (83.9-92.5) | ||
RT-PCR. real-time reverse transcriptase-polymerase chain reaction; LFI, lateral flow immunoassay; Sen, sensitivity; Sp, specificity; PPV, positive predictive values; NPV, negative predictive values.
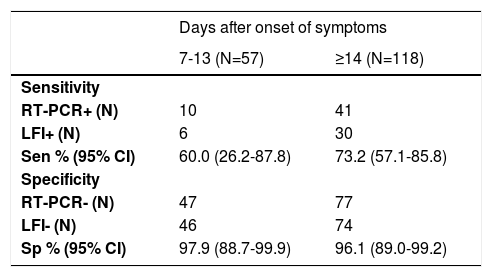
Sensitivity according to the two-time intervals from onset of symptoms was 60.0% (95%CI 26.2-87.8) and 73.2% (95%CI 57.1-85.8) at 7-13 and ≥14 days, respectively (Table 3). Specificity at 7-13 and ≥14 days of the onset of symptoms was 97.9% (95%CI 88.7-99.9) and 96.1% (95%CI 89.0-99.2) respectively (Table 3).
Sensitivity and specificity according to the days after onset of symptoms.
RT-PCR, real-time reverse transcriptase-polymerase chain reaction; LFI, lateral flow immunoassay; Sen, sensitivity; Sp, specificity.
The analysis to assess the test accuracy between infants (<2 years old) and older children (≥2 years old) did not show significant differences. LFI test overall sensitivity and specificity for those <2 years old (n=36, PCR+=10) were 70% (95%CI 34.8-93.3) and 100% (95%CI 86.8-100.0) respectively, while for children ≥2 years old (n=139, PCR+= 41) sensitivity was 70.7% (95%CI 54.5-83.9) and specificity 95.9% (95%CI 89.9-98.9).
DiscussionTo the authors' knowledge, this is the first study testing LFI accuracy in the pediatric population in a prospective study during the COVID-19 pandemic. The specificity found was over 96% regardless of days after the onset of symptoms. However, the sensitivity of LFI was low (73.2%), even lower during the second week after disease onset. This finding is in contrast with the commercial test description and illustrates the importance of clinical studies to validate any test in a specific population. Accuracy did not differ between infants and older children.
The authors found a lower sensitivity than in most other reports, but it is important to point out that the performance of these tests may change according to different manufacturers. Previous studies in adults have shown sensitivity numbers around 80%-90%.20–23 Further, most of these studies evaluated patients with more severe clinical courses and the possible lower levels of antibodies in milder disease may also account for such differences. The exact immunologic specificities between SARS-CoV-2 infected children and adults are still unclear and under investigation. Possible explanations may include a lower expression of angiotensin-converting enzyme 2 receptors, immature humoral and cellular immune responses, and different innate and adaptive responses in children, when compared to adults.24–26
The rapid point-of-care diagnosis was identified as a research priority by WHO R&D Blue Print expert group.27 Mean days of seroconversion for adult COVID-19 confirmed cases is around 9-12 days post onset.27,28 Some studies describe almost 100% of seroconversion in adult patients within 3 weeks of symptoms onset.29 Although there are no previous reports of the accuracy of serologic LFI in children and adolescents, studies about serology in this age group may be a proxy for the understanding of both tests. Serological diagnosis is especially important for patients who seek health care late for a molecular test.
The results of the present study suggest that a positive result may confirm the diagnosis, but a negative result is not sufficient to rule out the disease in children. In day-to-day practice, the seek for care usually occurs during the first week after the onset of signs and symptoms, when the sensitivity of the serological test is extremely low. Consequently, the LFI has no role in this scenario. If used in epidemiological surveys, the limitations in sensitivity must be considered for the interpretation of such data. It may be useful to evaluate clusters of cases, since a COVID-19 early diagnosis may allow confirmation of subsequent cases by epidemiological criteria. Furthermore, as serological tests are useful as diagnostic criteria for MIS-C, LFI may be helpful in the differential diagnosis of this syndrome at ERs.30 Despite these limitations, LFI testing is simple to perform and has a low cost, with potential use especially in low-resource or remote settings.
This study has some limitations. Firstly, 179 patients did not return for LFI testing. Pediatric patients with COVID-19 usually have a good prognosis and caregivers probably had the concern to return to a hospital setting for further testing when the clinical evolution was mild. Patients who did not return were younger, but the test performance was not different between age groups. A higher proportion of caucasian children did not undergo LFI testing, however, the authors believe that such differences would not change our overall results. Still, individuals who did not return could have a milder clinical course. Although controversial, mild disease has been related to lower levels of antibodies.31 Therefore, due to this subject loss in the study, the sensitivity found could be overestimated. Also, RT-PCR was used as the reference diagnostic method even with limitations such as the timing for collection and a sensitivity around 71%,32–34 which might have led to an underestimation in specificity and overestimation in sensitivity observed for the LFI test. Another point to address, the RT-PCR sample collection was performed 8 days or later after the onset of symptoms in 28 children, which could result in more false negative RT-PCR results. This might lead to underestimation of the specificity and overestimation of the LFI sensitivity. Finally, in the present study's sample, COVID-19 prevalence was around 30%. Interpretation of these results must be careful in different epidemiological scenarios, as accuracy (especially PPV and NPV) may change according to prevalence.
ConclusionDespite the high specificity (>96%) of LFI in a pediatric population, the sensitivity was lower than reports among adults. Moreover, the use of LFI should not be recommended during the first two weeks after the onset of symptoms. Further studies are necessary to assess the long-term performance of this test in children, as it is not known how long antibodies remain positive after an acute infection.
FundingThis work was supported by the Ministry of Health of Brazil, through the Institutional Development Program of the Brazilian National Health System (PROADI-SUS) in collaboration with Hospital Moinhos de Vento. The Ministry of Health of Brazil donated the rapid LFI tests used in the study, but had no role in study design; in the collection, analysis and interpretation of data; and in the decision to submit the article for publication.
The authors thank the Scientific Committee of the Research Support Nucleus (NAP) of Moinhos de Vento Hospital for technical-scientific consultancies. The authors thank Matias Segelis Vieira, statistician who supported the data analysis, the field workers, laboratory team, and the staff from Hospital Moinhos de Vento, and from Hospital Restinga e Extremo Sul: Aline Andrea da Cunha, Sidiclei Machado Carvalho, Fabio Jose Rockenbach, Kelly Viegas Antunes, Marcelo da Silva Ferreira, Rafael Garcia Trindade, Thayna Silva Lino, Tiago da Silva Silvano, Adriana da Silva Silveira, Alceu Kuckoski, Ana Paula da Silva Lopes, Andreia Escobar da Costa, Clarice Cardoso Machado, Erica Vieira da Silva, Evelin Inácia da Silva, Luciana Rodrigues Ribeiro, Marcely Mayr da Costa, Morgana Thais Carollo Fernandes, Rafael da Silva Cassafuz.
These authors contributed equally to this manuscript.
COVIDa study group: Camila Dietrich, Tiago Fazolo, Fernando Rovedder Boita, Fernanda Lutz Tolves, Jaina da Costa Pereira, Adriana Isabel Rohden, Glaucia Fragoso Hohenberger, Thainá Dias Luft, Shirlei Villanova Ribeiro, Catia Moreira Guterres, Caroline Cabral Robinson, Débora Vacaro Fogazzi, Eliana Marcia da Ros Wendland, Regis Goulart Rosa, Maristênia Machado Araújo, Raul Correia da Silva, Ana Carolina Monteiro da Rocha, Tássia Rolim Camargo, Giovana Petracco de Miranda, William Jones Dartora, Camila Bonalume Dall'Aqua, Thaís Jacobsen Duarte, Madelaine Correa de Oliveira, Emerson Silveira de Brito, Thayane Martins Dornelles, Ana Paula dos Santos, Gisele Alcina Nader Bastos, Denise Arakaki-Sanchez, Leonardo Araújo Pinto, Maicon Falavigna, Patricia Bartholomay Oliveira e Francieli Fontana Sutile Tardetti Fantinato.