To contribute to a better understanding of the maternal genetic mechanisms that influence obstetric outcomes and that are involved in maternal and child health, this study aimed to evaluate the association between maternal genetic variants and the offspring birth weight by analyzing single-nucleotide polymorphisms (SNPs) in genes related to glucose homeostasis.
MethodsThree polymorphisms were analyzed (GCK rs1799884, TCF7L2 rs7903146 and LEPR rs1137101) in 250 pregnant women who participated in a Brazilian prospective cohort study. Genotyping was performed by Real-Time Polymerase Chain Reaction (qPCR) using pre-designed TaqMan® SNP genotyping assays. Vitamin D dosage was performed by chemiluminescence. Variance, Pearson's chi-square test and multiple linear regression were used for the statistical analysis.
ResultsIt was possible to verify a significant association between birth weight and maternal GCK rs1799884 when obstetric outcomes, clinical and anthropometric characteristics were taken into consideration. The children of homozygous women for the minor allele GCK rs1799884 presented lower birth weight (β = -335.25, 95% CI = -669.39; -1.17, p = 0.04). Furthermore, a direct link between a leptin receptor variant and gestational duration was found (p = 0.037).
ConclusionThe variant GCK rs1799884 (mm) was associated with a reduction in newborn weight in the miscegenated Brazilian population.
A clear understanding of both environmental and genetic determinants of fetal growth is vital to determine long-term consequences of low birth weight and clinical management of newborns.1,2 Birth weight is one of the main parameters used to evaluate newborn and infant health and survival and to predict future complications in adulthood.3,4 There are pieces of evidence linking birth weight with the development of chronic diseases in adult life, such as diabetes, hypertension, cardiovascular disease, and other illnesses.1,5
During pregnancy, maternal metabolism is a major determinant of the intrauterine environment and fetal outcomes.6 During this period, metabolic adaptations are essentials to meet maternal physiological demands, as well as fetal adequate growth and development.7 Among the main alterations observed in pregnant women, glucose metabolism is noteworthy, once it is the primary source of fetal energy.6
Maternal glucose is a key element of fetal growth, and therefore genetic variants in the mother that impact glucose levels may modify fetal development indirectly by changing maternal glycemia, which stimulates fetus insulin production.4,8 This modification can alter birth weight because insulin is an important intrauterine growth factor.8,9 Owing to the significant role of maternal glucose concentration as a determining factor of offspring birth weight,2,9 genes that alter glucose homeostasis are good candidates for fetal growth biological markers, due to modification of insulin action and/or release.8
The Glucokinase enzyme, encoded by the gene with the same name (GCK), is one of the main regulators of fasting glucose concentrations.9 It is responsible for catalyzing the first rate-limiting reaction in the glycolysis pathway and controls glucose-stimulated insulin secretion from pancreatic β-cells and glucose metabolism in the liver.10,11
The TCF7L2 gene encodes the Transcription Factor 7-Like-2, which plays a central role in coordinating the expression and processing of insulin.12 Furthermore, it is known to be associated with Type 2 diabetes and gestational diabetes, as well as to be linked to fasting glucose levels in the general population.6,13
Leptin, also known as adipose tissue peptide hormone, is involved in the regulation of body fat and metabolism, besides having several other complex actions on insulin-sensitive tissues and endocrine pancreas hormones, which appear to contribute to glucose homeostasis.14–16 Leptin action occurs through its receptor that is encoded by the LEPR gene. Variations in leptin receptors may play a role in maternal body weight during pregnancy and fetal growth.14
In the present study, the authors evaluated the influence of maternal genetic variants on the offspring birth weight by analyzing polymorphisms in genes related to glucose homeostasis (GCK rs1799884, TCF7L2 rs7903146 and LEPR rs1137101) in order to contribute to a better understanding of the maternal genetic mechanisms that influence obstetric outcomes and that are involved in maternal and child health.
Material and methodsStudy design and sampleThis is a prospective cohort study with pregnant women of the Maternal and Child Health Research Center (NISAMI) from Santo Antônio de Jesus, Bahia, Northeast of Brazil. The NISAMI cohort was developed in the urban area of the city and aimed to investigate determinants of maternal and infant health.
The study population consisted of 250 pregnant women from the NISAMI cohort. They were clinically healthy women, 18 and above years old, resident and domiciled in the urban area of Northeast of Brazil, who attended the service of prenatal care of the public health system (SUS). Data were collected between 2013 and 2015.
Exclusion and inclusion criteriaThe eligibility criteria to participate in this study were a gestational age of 34 or fewer weeks at the time of the first contact. Exclusion criteria were women with multiple gestations; HIV positive; with contagious, immunological and metabolic diseases; pregnant women who were vegetarian or vegan (did not consume any animal food); besides those who suffered a miscarriage during the cohort follow-up.
Data collection and definition of variablesInformation related to the socioeconomic and demographic conditions, lifestyle, health, and obstetric data were collected by trained interviewers and recorded in a standardized questionnaire.
In the first stage of the cohort (baseline) when a pregnant woman was included in the study, the woman's pre-gestational weight was collected from the pregnancy chart. When not available, the weight provided by the woman was taken during the interview.17 Maternal height was measured by the health team at the clinic, who were trained according to the recommendations of Lohman et al. (1998).18 After these procedures, a date was scheduled for blood collection.
Gestational age was calculated based on the date of the last menstrual cycle, available on the pregnant woman's chart, or the gestational age was recorded from the first ultrasound, which was performed by the end of the first trimester.17
Blood collection for DNA extraction and vitamin D DosingBlood collection was performed between 7:00 and 9:00 a.m. after at least 8 hours of fasting at the Research Center for Maternal and Child Health of the municipality. Fasting time was based on other biochemical analyses performed with pregnant women. Blood was centrifuged for 15 min at 2000 rpm in a UV light protected environment for serum separation.
Vitamin D dosage was performed by the quantitative determination method, based on chemiluminescence. A 25(OH)D dosing kit was used (DiaSorin®), with an intra-assay CV of 8.4–12.5% and an inter-assay CV of 8.6–11.0%.
Molecular analysisGenomic DNA was isolated using Qiagen® FlexiGene DNA Kit (Qiagen, CA, USA) according to the manufacturer's recommendations. After extraction, DNA concentration and purity were determined by the Thermo Scientific NanoDropTM 2000 Micro-volume Spectrophotometer (Thermo Fisher Scientific Inc., Wilmington, DE, USA). DNA extraction and measurement of concentration and purity were carried out at the Universidade Federal do Recôncavo da Bahia Human Genetics Laboratory.
SNPs selection was based on the relationship with glucose homeostasis. GCK rs1799884, TCF7L2 rs7903146 and LEPR rs1137101 polymorphisms were genotyped for all samples at the Center of Human and Molecular Genetics (NGHM – Núcleo de Genética Humana e Molecular) of the Universidade Federal do Espírito Santo, Vitória, ES, Brazil. Genotyping was performed by Real-Time Polymerase Chain Reaction (qPCR) using pre-designed TaqMan® SNP genotyping assays (Applied Biosystems, Foster City, CA, USA). The Real-Time PCR equipment was the Applied Biosystems 7500 Fast Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). Allelic discrimination was made using software 7500, version 2.0.6 from the Real-Time PCR equipment. All reactions were performed according to the manufacturer's recommendations and known positive and negative controls were used.
Neonatal outcomesBetween September 2013 and December 2015, pregnant women were monitored until childbirth. Naked childbirth weight was measured using a digital pediatric scale, Welmy® brand, with a capacity of 15kg and a 10g interval. The anthropometric measurement was performed in duplicate, and a maximum variation of 10 g weight was accepted. When different values were obtained, a third measurement was performed. The final measurement was the closest measurement mean value.19
Data related to children's birth were collected at the Department of Epidemiological Surveillance of the Municipal Health Department. Home visits were performed at the end of the study for pregnant women whose gestational outcome data were not found.
Statistical analysisDescriptive analyses were used to characterize the sample using absolute and relative frequencies to describe categorical variables. For discrete and continuous variables, mean and standard deviation (SD) were used. Analysis of variance (One-Way ANOVA) and Bonferroni test was used to compare continuous variables (birth weight averages, pre-pregnancy maternal body mass index (BMI) and gestational age) according to each polymorphism genotypes. Pearson's chi-square test was used to verify the relationship between categorical variables and polymorphism genotypes. Simple and multiple linear regression analyses were performed to verify the influence of maternal polymorphisms on newborn's weight, considering baby sex, pre-pregnancy maternal BMI, gestational age, vitamin D serum concentration, planned pregnancy, maternal education (< 12 years), and race (brown/black). Linear regression model assumptions were analyzed. Normality was assessed by analysis of the adjusted residual and linearity and homoscedasticity by graphical inspection. Collinearity was evaluated through factor inflation variance. All assumptions were met for all defined models. Data were analyzed by the Stata software version 14. The significance threshold level of p < 0.05 was adopted.
Genotype distribution was tested for the Hardy-Weinberg equilibrium using Arlequin software version 3.5.2.2. Values of p < 0.05 were considered statistically significant.
Ethics approvalThe study was approved by the Research Ethics Committee involving Human Beings, of the Universidade Federal do Recôncavo da Bahia (UFRB), number: 241.225 dated 04/09/2013. All study proceedings have been carried out in accordance with The Code of Ethics of the World Medical Association (Declaration of Helsinki) for experiments involving humans. Informed consent was obtained for experimentation with human subjects and the privacy rights of human subjects were observed.
ResultsParticipant characteristicsThe main subject characteristics included in this study (mother and newborn) are shown in Table 1. The final sample consisted of 250 women, 84% (n=210) black/brown, with mean age of 27.24 years (SD = 6.01), pre-pregnancy BMI of 24.17 Kg/m² (SD= 4.90) and concentrations of 25-hydroxyvitamin D (25(OH)D) of 72.13 nmol/l (SD= 30.47). Children had a mean birth weight of 3324 g (SD = 494.60) and were born with a mean gestational age of 38.59 weeks (SD = 1.96).
Main characteristics of NISAMI cohort (n = 250).a
| Characteristics | Mean (SD) or n (%)b |
|---|---|
| Maternal | |
| Age (years) | 27.24 (6.01) |
| Weight (kg) | 67.17 (12.82) |
| Height (m) | 1.61 (0.06) |
| Pre-pregnancy BMI (kg/m²) | 24.17 (4.90) |
| Serum Vitamin D (nmol/l) | 72.13 (30.47) |
| Race | |
| Black | 105 (42.00%) |
| Brown | 105 (42.00%) |
| White | 25 (10.00%) |
| Other | 15 (6.00%) |
| Planned pregnancy | 93 (37.05%) |
| Education (< 12 years) | 179 (71.31%) |
| Obstetric outcomes | |
| Sex of the baby | |
| Female | 134 (53.60%) |
| Male | 116 (46.40%) |
| Birth weight (g) | 3,324 (494.60) |
| Low (< 2,500) | 11 (4.40%) |
| Insufficient (2,500 to 2,999) | 49 (19.60%) |
| Adequate (3,000 to 3,999) | 171 (68.40%) |
| Excessive (≥ 4,000) | 19 (7.60%) |
| Gestational age (weeks) | 38.59 (1.96) |
| Preterm (< 37) | 33 (13.20%) |
| Term (37 to < 42) | 208 (83.20%) |
| Post-term (≥42) | 9 (3.60%) |
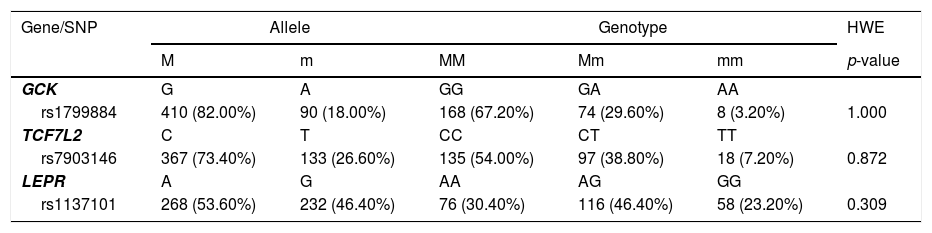
Genotype calls were successful for all 250 samples. Maternal GCK rs1799884, TCF7L2 rs7903146 and LEPR rs1137101 genotype distribution and allele frequencies are presented in Table 2. Minor alleles are 18.00%, 26.60% and 46.40%, respectively. No deviation from Hardy-Weinberg equilibrium was observed (p > 0.05).
Allele frequencies and genotype distribution.
M, major allele; m, minor allele; MM, homozygous for the major allele; Mm, heterozygous; mm, homozygous for the minor allele; HWE, Hardy-Weinberg Equilibrium.
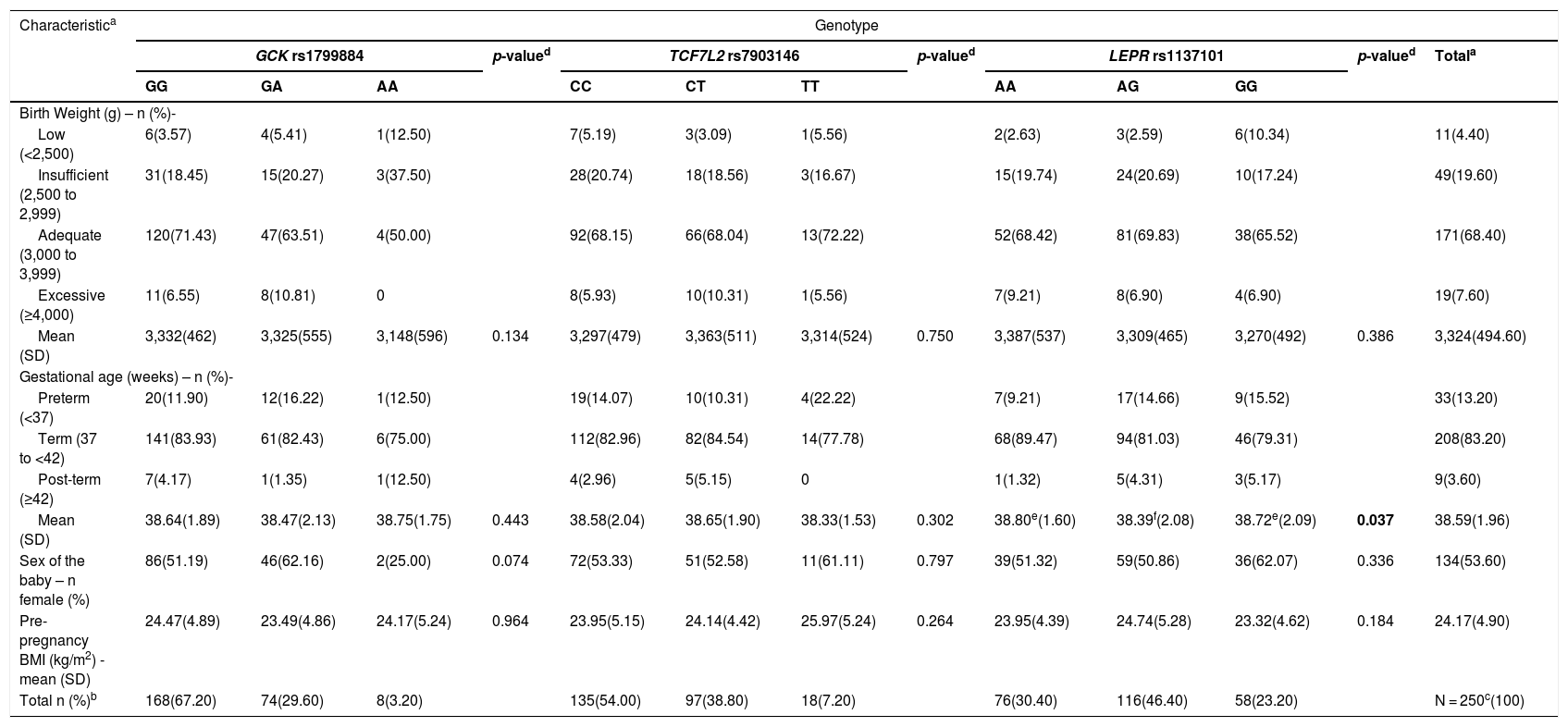
Maternal genotypes were related to obstetric outcomes and maternal anthropometric parameters, as shown in Table 3. No significant difference between birth weight classification (low, insufficient, adequate, and excessive) and gestational age classification (preterm, term and post-term) with polymorphisms were found. Despite observed tendencies, no significant association between maternal genetic variants and mean birth weight was found. However, a significant correlation between LEPR rs1137101 and mean gestational age was observed (p = 0.037), since patients who were heterozygous LEPR rs1137101 were born at a significantly younger gestational age.
Maternal genotype correlation with obstetric outcomes and anthropometric parameters.
| Characteristica | Genotype | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GCK rs1799884 | p-valued | TCF7L2 rs7903146 | p-valued | LEPR rs1137101 | p-valued | Totala | |||||||
| GG | GA | AA | CC | CT | TT | AA | AG | GG | |||||
| Birth Weight (g) – n (%)- | |||||||||||||
| Low (<2,500) | 6(3.57) | 4(5.41) | 1(12.50) | 7(5.19) | 3(3.09) | 1(5.56) | 2(2.63) | 3(2.59) | 6(10.34) | 11(4.40) | |||
| Insufficient (2,500 to 2,999) | 31(18.45) | 15(20.27) | 3(37.50) | 28(20.74) | 18(18.56) | 3(16.67) | 15(19.74) | 24(20.69) | 10(17.24) | 49(19.60) | |||
| Adequate (3,000 to 3,999) | 120(71.43) | 47(63.51) | 4(50.00) | 92(68.15) | 66(68.04) | 13(72.22) | 52(68.42) | 81(69.83) | 38(65.52) | 171(68.40) | |||
| Excessive (≥4,000) | 11(6.55) | 8(10.81) | 0 | 8(5.93) | 10(10.31) | 1(5.56) | 7(9.21) | 8(6.90) | 4(6.90) | 19(7.60) | |||
| Mean (SD) | 3,332(462) | 3,325(555) | 3,148(596) | 0.134 | 3,297(479) | 3,363(511) | 3,314(524) | 0.750 | 3,387(537) | 3,309(465) | 3,270(492) | 0.386 | 3,324(494.60) |
| Gestational age (weeks) – n (%)- | |||||||||||||
| Preterm (<37) | 20(11.90) | 12(16.22) | 1(12.50) | 19(14.07) | 10(10.31) | 4(22.22) | 7(9.21) | 17(14.66) | 9(15.52) | 33(13.20) | |||
| Term (37 to <42) | 141(83.93) | 61(82.43) | 6(75.00) | 112(82.96) | 82(84.54) | 14(77.78) | 68(89.47) | 94(81.03) | 46(79.31) | 208(83.20) | |||
| Post-term (≥42) | 7(4.17) | 1(1.35) | 1(12.50) | 4(2.96) | 5(5.15) | 0 | 1(1.32) | 5(4.31) | 3(5.17) | 9(3.60) | |||
| Mean (SD) | 38.64(1.89) | 38.47(2.13) | 38.75(1.75) | 0.443 | 38.58(2.04) | 38.65(1.90) | 38.33(1.53) | 0.302 | 38.80e(1.60) | 38.39f(2.08) | 38.72e(2.09) | 0.037 | 38.59(1.96) |
| Sex of the baby – n female (%) | 86(51.19) | 46(62.16) | 2(25.00) | 0.074 | 72(53.33) | 51(52.58) | 11(61.11) | 0.797 | 39(51.32) | 59(50.86) | 36(62.07) | 0.336 | 134(53.60) |
| Pre-pregnancy BMI (kg/m2) - mean (SD) | 24.47(4.89) | 23.49(4.86) | 24.17(5.24) | 0.964 | 23.95(5.15) | 24.14(4.42) | 25.97(5.24) | 0.264 | 23.95(4.39) | 24.74(5.28) | 23.32(4.62) | 0.184 | 24.17(4.90) |
| Total n (%)b | 168(67.20) | 74(29.60) | 8(3.20) | 135(54.00) | 97(38.80) | 18(7.20) | 76(30.40) | 116(46.40) | 58(23.20) | N = 250c(100) | |||
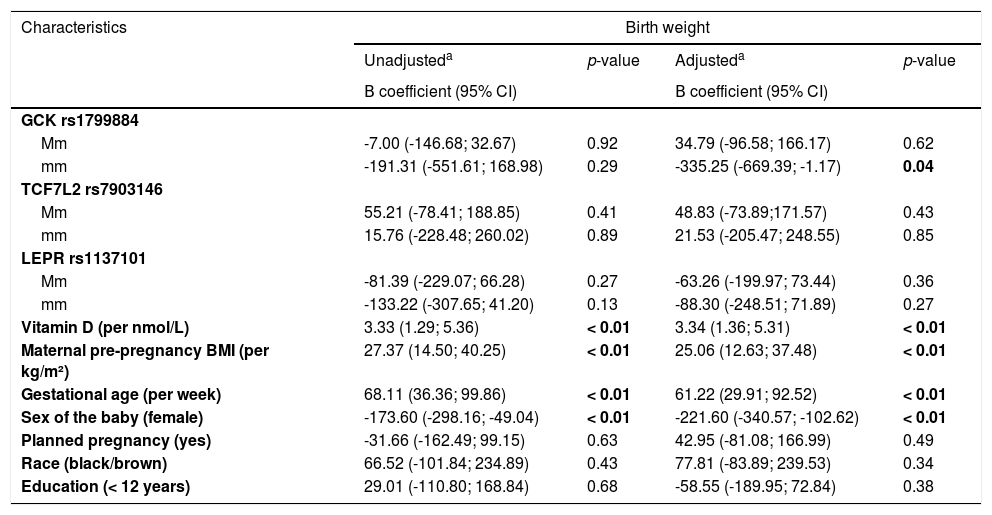
To verify the influence of maternal genetic variants on newborn's weight considering other characteristics, simple and multiple linear regression analyses were performed and have found a significant association (p < 0.01) between birth weight and covariables baby sex, pre-pregnancy maternal BMI, gestational age, and vitamin D serum concentration (Table 4). Furthermore, the adjusted model for the association between birth weight and homozygous mothers for the minor allele GCK rs1799884 showed a 335g (95% CI = -669.39; -1.17, p = 0.04) decrease in birth weight compared to the MM (homozygous for the major allele) group.
Influence of maternal genetics on birth weight, considering obstetric outcomes, clinical and anthropometric characteristics.
| Characteristics | Birth weight | |||
|---|---|---|---|---|
| Unadjusteda | p-value | Adjusteda | p-value | |
| Β coefficient (95% CI) | Β coefficient (95% CI) | |||
| GCK rs1799884 | ||||
| Mm | -7.00 (-146.68; 32.67) | 0.92 | 34.79 (-96.58; 166.17) | 0.62 |
| mm | -191.31 (-551.61; 168.98) | 0.29 | -335.25 (-669.39; -1.17) | 0.04 |
| TCF7L2 rs7903146 | ||||
| Mm | 55.21 (-78.41; 188.85) | 0.41 | 48.83 (-73.89;171.57) | 0.43 |
| mm | 15.76 (-228.48; 260.02) | 0.89 | 21.53 (-205.47; 248.55) | 0.85 |
| LEPR rs1137101 | ||||
| Mm | -81.39 (-229.07; 66.28) | 0.27 | -63.26 (-199.97; 73.44) | 0.36 |
| mm | -133.22 (-307.65; 41.20) | 0.13 | -88.30 (-248.51; 71.89) | 0.27 |
| Vitamin D (per nmol/L) | 3.33 (1.29; 5.36) | < 0.01 | 3.34 (1.36; 5.31) | < 0.01 |
| Maternal pre-pregnancy BMI (per kg/m²) | 27.37 (14.50; 40.25) | < 0.01 | 25.06 (12.63; 37.48) | < 0.01 |
| Gestational age (per week) | 68.11 (36.36; 99.86) | < 0.01 | 61.22 (29.91; 92.52) | < 0.01 |
| Sex of the baby (female) | -173.60 (-298.16; -49.04) | < 0.01 | -221.60 (-340.57; -102.62) | < 0.01 |
| Planned pregnancy (yes) | -31.66 (-162.49; 99.15) | 0.63 | 42.95 (-81.08; 166.99) | 0.49 |
| Race (black/brown) | 66.52 (-101.84; 234.89) | 0.43 | 77.81 (-83.89; 239.53) | 0.34 |
| Education (< 12 years) | 29.01 (-110.80; 168.84) | 0.68 | -58.55 (-189.95; 72.84) | 0.38 |
Mm, heterozygous; mm, homozygous for the minor allele; MM, homozygous for the major allele.
In this study, the authors analyzed the association between maternal variants in genes related to glucose homeostasis with birth weight, as well as with obstetric outcomes, clinical and anthropometric characteristics. An unadjusted association of genetic variants with birth weight was not found, but after adjustment for covariables, the authors detected a significant relation between maternal GCK rs1799884 and lower birth weight.
Worldwide studies have investigated the association of maternal polymorphisms and birth weight, but they are vastly based on European Caucasian populations. The lack of data in different ethnic groups highlights the need to study the genetic influence on newborn weight in diverse ethnic groups.2 The present study's results represent a unique controlled prospective cohort, made of predominantly black/brown Brazilian participants.
The investigation of GCK rs1799884 in pregnant European Caucasian women verified an association of this polymorphism and newborn weight. It has been found that the presence of the minor allele increases fetal growth due to the increase in maternal glucose levels, which reflects in birth weight.9,10 In contrast, the present study's results revealed a minor allele trend to decrease offspring birth weight.
TCF7L2 rs7903146 is an important polymorphism associated with diabetes type 2.6,13 A white European subject meta-analysis revealed that maternal TCF7L2 polymorphic allele presence increased newborn birth weight,8 which was attributed to the known effect of TCF7L2 rs7903146 on reducing insulin secretion, resulting in higher maternal glycemia. In the present study, the authors identified that newborns of variant allele mothers have an increased birth weight, but the association was not significant.
The authors have shown that there is no correlation between LEPR rs1137101 and birth weight. Similar results were found in populations from the UK/Irish and South Asia that were analyzed for the influence of maternal genetic variants on maternal BMI, birth weight and gestational length and did not find any significant associations.14 Interestingly, while that study observed an increase in birth weight, the present study showed a decrease in newborn's weight, but both were not statistically significant. Furthermore, the authors have found a significant association between LEPR rs1137101 and gestational age, in contrast to the UK/Irish-South Asian study.
The present study results suggest that factors like newborn sex, maternal BMI, gestational age, and vitamin D are linked to offspring birth weight in the Brazilian population. The authors analyzed the influence of maternal genetic variants on newborn's weight considering other characteristics, and the authors have found a significant association between birth weight and variables baby sex, pre-pregnancy maternal BMI, gestational age, and serum Vitamin D concentration. The influence of these covariables on birth weight is in accordance with what is shown in the literature, i.e.: female fetuses showed less weight gain than male fetuses,20 maternal BMI had a directly proportional influence on the newborn's weight,21 the gestational age was a strong contributor to birth weight,22 and that inadequate vitamin D during pregnancy is associated with low birth weight.23 Also, after adjusting for those covariables, the authors observed that mothers who are homozygous for the minor allele GCK rs1799884 have newborns with a significant decrease in birth weight.
A controlled prospective cohort study with standardized data collection that adopted strategies to avoid data collection bias were the positive aspects of this study. The limitation was mainly sample number when compared to larger European studies. Furthermore, the present study's results could be confounded with other variables not analyzed, such as blood glucose and its metabolism. However, these aspects did not compromise the validity of the present study, considering that the data from this investigation adopted strategies to avoid data collection bias risk.
The present study contributes to the understanding of genetic influences on birth weight from different ethnical groups, other than European populations. The Brazilian population has considerable ethnic miscegenation and has been classified as one of the most heterogeneous populations in the world,24 highlighting the importance of the present study and possibly explaining some differences in the present study's results.
In conclusion, the present study shows that maternal GCK 1799884 minor allele influences lower birth weight. Furthermore, maternal TCF7L2 rs7903146 and LEPR rs1137101 do not appear to have a direct impact on birth weight. To the authors’ knowledge, this work is a pioneer in investigating the relationship between maternal genetic variants related to glucose metabolism and birth weight in Brazil. Further studies are required to test more genetic variants, in order to better assist pregnant women and children in ensuring the well-being of mother and baby.
This study was supported by Fundação de Amparo à Pesquisa do Espírito Santo (FAPES), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES). The authors thank all participants and collaborators.
1Study conducted at the Universidade Federal do Recôncavo da Bahia, Santo Antônio de Jesus, Bahia, Brazil.