This narrative, non-systematic review provides an update on the genetic aspects of the SARS-CoV-2 virus and its interactions with the human genome within the context of COVID-19. Although the main focus is on the etiology of this new disease, the genetics of SARS-CoV-2 impacts prevention, diagnosis, prognosis, and the development of therapies.
Data sourceA literature search was conducted on MEDLINE, BioRxiv, and SciELO, as well as a manual search on the internet (mainly in 2019 and 2020) using the keywords “COVID-19,” “SARS-CoV-2,” “coronavirus,” “genetics,” “molecular,” “mutation,” “vaccine,” “Brazil,” “Brasil,” and combinations of these terms. The keywords “Brazil” and “Brasil” were used to find publications that were specific to the Brazilian population’s molecular epidemiology data. Articles most relevant to the scope were selected non-systematically.
Data synthesisA number of publications illustrate an expanding knowledge on the genetics and genomics of SARS-CoV-2 and its implications for understanding COVID-19.
ConclusionsKnowledge of the SARS-CoV-2 genome sequence permits an in-depth investigation of the role its proteins play in the pathophysiology of COVID-19, which in turn will be enormously valuable for understanding the evolutionary, clinical, and epidemiological aspects of this disease and focusing on prevention and treatment.
“…if you know your enemies and know yourself, you will not be imperiled in a hundred battles.” Sun Tzu, The Art of War
On December 31, 2019, four cases of pneumonia of unknown etiology in Wuhan (Hubei province) were reported to the Chinese office of the World Health Organization.1 In record time, on January 12, 2020, Chinese researchers shared the genetic sequence of the etiological agent,2 the virus that would be called SARS-CoV-23 and caused what came to be known as COVID-19.4 On February 26, 2020, Brazil’s first case of COVID-19 was confirmed in a 61-year-old man who had recently traveled to the Lombardy region of Italy.5 A day later, the genome of the SARS-CoV-2 virus from this patient was sequenced, a great accomplishment for Brazilian science.6 The virus has a unique sequence of bases that differs from other species and is genetically different enough from SARS-CoV-1 (genetic similarity of approximately 79%) and MERS-CoV (50%) to be considered a new virus. Phylogenetic analyses placed it in the genus Betacoronavirus, within the subgenus Sarbecovirus of the Coronaviridae family.7 The main goal of this narrative, non-systematic review is to provide an update on the genetic aspects of the SARS-CoV-2 virus and its interactions with the human genome within the context of COVID-19. Although the main focus is on the etiology of this new disease, the genetics of SARS-CoV-2 impacts prevention, diagnosis, prognosis, and the development of therapies.
MethodThe published scientific literature regarding “Genetics of COVID-19” was searched in a variety of databases, including MEDLINE, BioRxiv, and SciELO, as well as a manual search on the internet (mainly in 2019 and 2020), to ensure that the majority of relevant studies had been identified. The author called out several keywords (such as “COVID-19,” “SARS-CoV-2,” “coronavirus,” “genetics,” “molecular,” “mutation,” “vaccine,” “Brazil,” “Brasil”) so others can identify the work during database searches. The keywords “Brazil” and “Brasil” were used to find publications that were specific to the Brazilian population’s molecular epidemiology data. After the search was complete and all duplicates were thrown out, the author reviewed the abstracts of the remaining articles to ensure that they address our review question. The thirty-seven articles most relevant to the scope were non-systematically selected. The author summarized and synthesized the findings from the articles found and integrated them into the writing as appropriate.
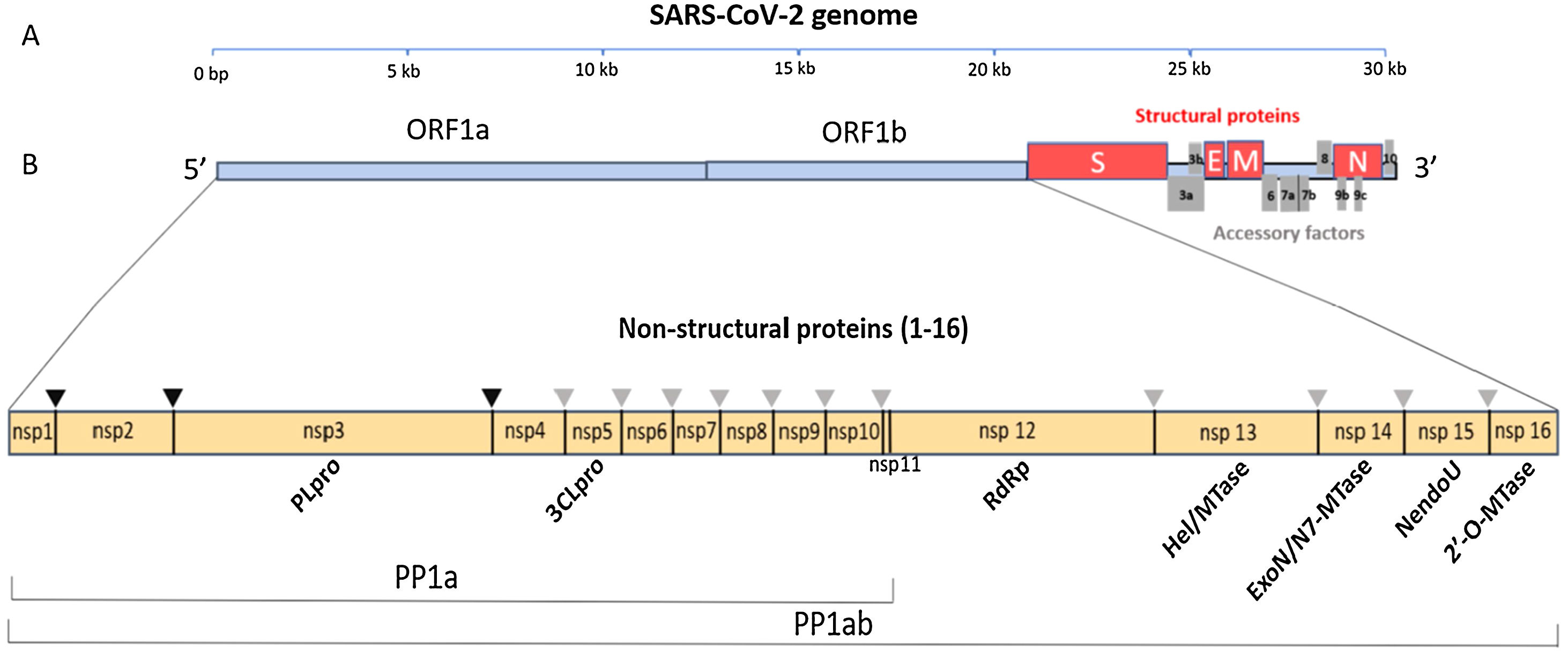
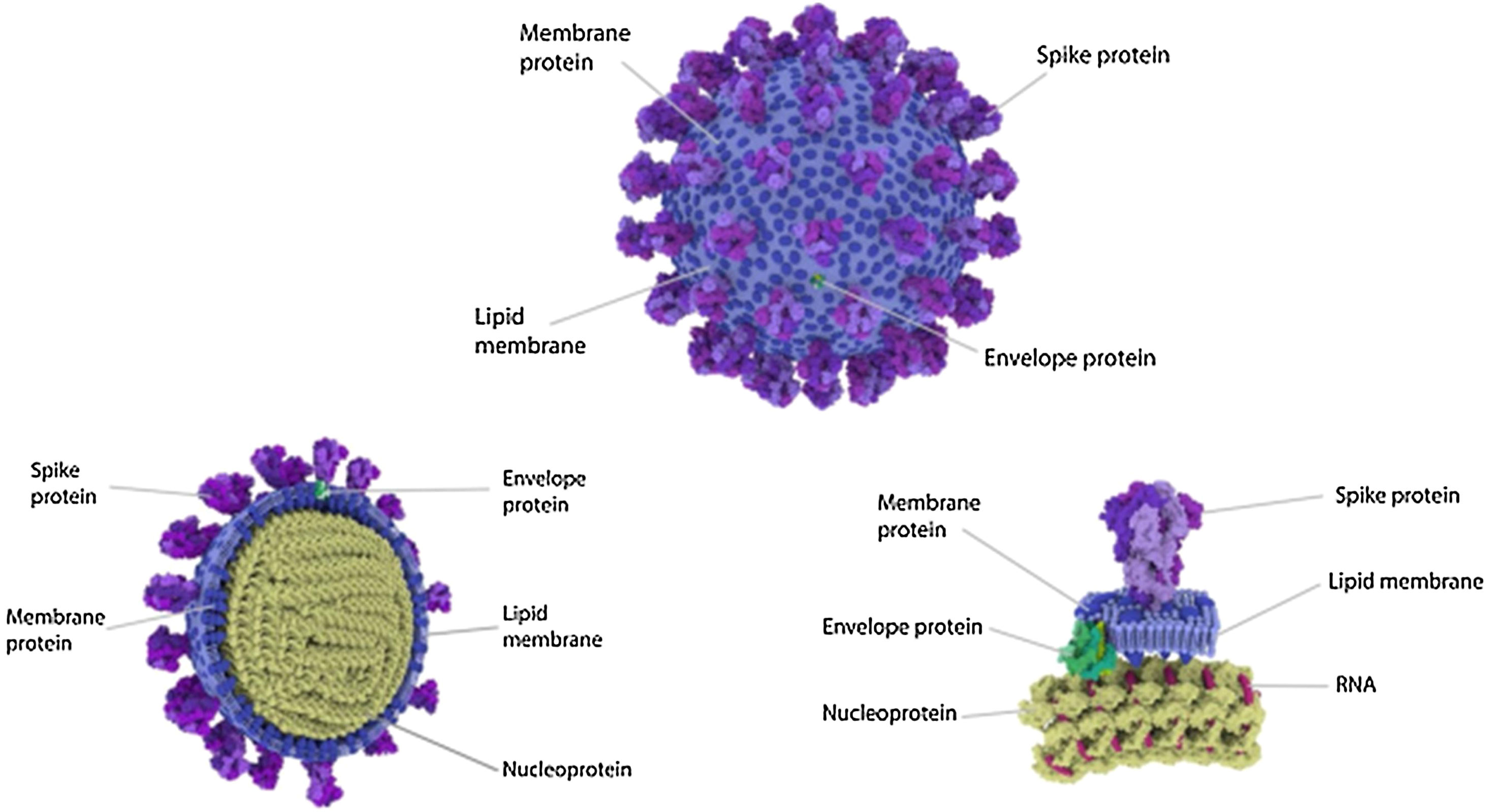
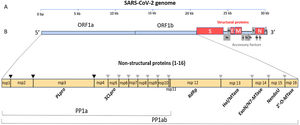
Data synthesisThe genome and proteome of SARS-CoV-2Like other coronaviruses, the genome of SARS-CoV-2 is composed of a single strand of RNA with a positive strand (ready for translation and consequent synthesis of its proteins). The genome is considered large, with 29,903 base pairs. There are at least 50 different sites where translation can begin (open reading frames – ORFs). These ORFs are each of the RNA sequences understood to include a start codon (AUG), a stop codon (UAG, UAA, or UGA), and the codons between them. This variable origin of transcription sequences allows the SARS-CoV-2 virus to encode for around 50 proteins that have non-structural, structural, and accessory functions.7,8 The initial two-thirds of the RNA sequence encode the two main transcriptional units, ORF1a and ORF1ab; these units encode two polyproteins (PP1a and PP1ab, respectively). The larger unit, PP1ab, contains ORFs for at least 16 non-structural proteins (Nsp1−16). The non-structural proteins have various functions in biological phenomena that are important for the virus such as replication, correction of replication errors (“proofreading”), translation, suppression of host proteins, immune response blockage, and RNA stabilization.8 The final third of the RNA encodes proteins that define the structure of SARS-CoV-2 as well as accessory proteins. Accessory genes are distributed among the genes that encode structural proteins and the 3' end of the genome and contain at least nine ORFs for accessory proteins; these proteins are not significant for viral replication but play an important role in interactions between the virus and host, including modulating and blocking the production of pro-inflammatory cytokines. Finally, three proteins that structure the virus, known as the spike (S), membrane (M), and envelope (E), are embedded in the outer membrane and give the virus its distinct shape and structure. Inside the virus particle, the RNA is tightly coiled and coated with a fourth structural protein, nucleocapsid (N), which protects its genetic material.7,8 New data shows a high-resolution map of the SARS-CoV-2 coding regions, allowing to accurately quantify the expression of canonical viral ORFs and to identify 23 unannotated viral ORFs. The new ORFs identified may serve as novel accessory proteins or as regulatory units controlling the balanced production of different viral proteins.7,8 So, it is possible that the SARS-CoV-2 30-thousand-base-pairs genome actually codes for 50 and not 27 proteins (Figs. 1 and 2).9
The SARS-CoV-2 genome has many ORFs and encodes as far as 50 non-structural, structural, and accessory proteins. Source: Romano et al.7.
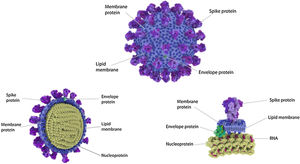
Four Structural Proteins in SARS-CoV-2. Spike (S), membrane (M), and envelope (E) are embedded in this outer membrane, providing the distinct shape and structure of the virus. Within the virus particle, the RNA is tightly coiled and coated with a fourth protein, nucleocapsid (N). Source: Slater.9.
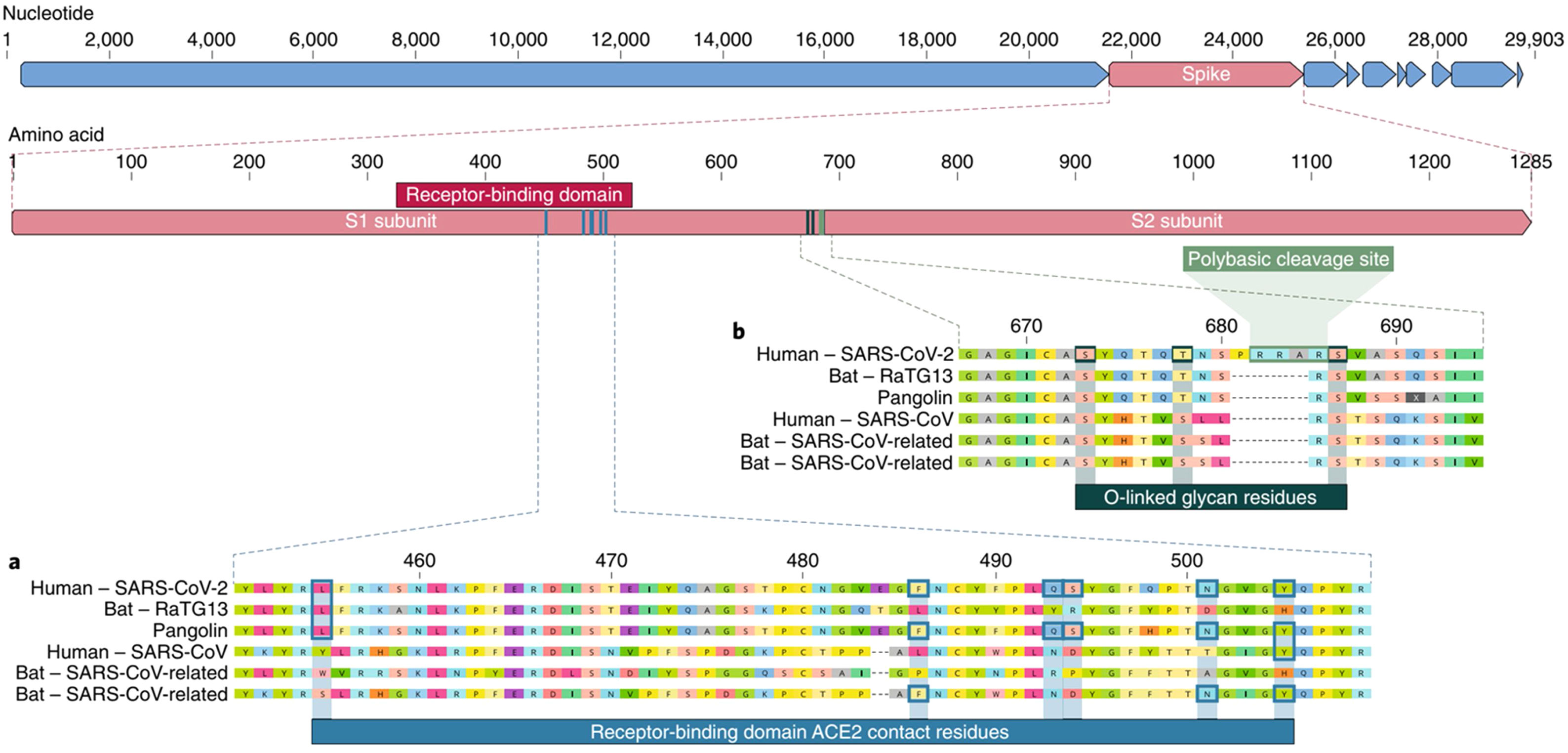
The spike protein (S) is encoded by 3831 base pairs of the SARS-CoV-2 RNA. Of the four structural proteins, this one selects which type of cell the SARS-CoV-2 will infect. The spike connects the virus and the human cell and causes the viral membrane to fuse with the cytoplasmic membrane of the human cell, so that the viral RNA can be injected into the human cell. Recognition of the membrane receptors in the cells to be infected is the mechanism by which tropism occurs. The spike, through its receptor-binding domain (RBD), recognizes the membrane receptor of angiotensin-converting enzyme 2 (ACE2), a protein expressed mainly in the lungs, heart, kidneys, and intestine. By selecting ACE2 as a target, the virus also selects the main tissues it will infect. Then, human transmembrane protease serine 2 (TMPRSS2) cleaves and activates the spike protein, which through its fusion peptide fuses the viral membrane with the membrane of the target cell, permitting injection of the SARS-CoV-2 RNA into the human cell.10 Inside the cell, the human protein synthesis apparatus (including the ribosomes, endoplasmic reticulum, and Golgi complex) is commandeered and used to translate the proteins of SARS-CoV-2, forming countless new virions that equip themselves to invade new cells before destroying the cell they have taken over. One of the main stages in this process is unique to SARS-CoV-2 and consists of pre-activation of a spike cleavage site by furin, a human protease present in the Golgi apparatus.11 Furin is significantly expressed in lung cells, and enveloped viruses take advantage of this fact to pre-activate their glycoproteins. One event in the pre-activation of the spike protein (pre-proteolysis) takes place in the Golgi complex of infected and virus-producing cells, since the Golgi contains furins activated by the local pH. This pre-activation by furin seems to grant better selectivity and tropism to the spike with relation to ACE2, allowing it to enter cells that have low expression of other proteases such as TMPRSS2 and lysosomal cathepsins. When the RBD of the spike connects to the host’s ACE2 receptor, the host proteases (particularly TMPRSS2 and lysosomal cathepsins, along with furin) cleave and activate the “pre-activated” spike protein at the S1/S2 junction, where a cleavage site recognized by the proteases is present. Activation of this site exposes a second protease cleavage site (S2). The spike protein must be cleaved sequentially at both sites, S1/S2 and S2, to be effectively activated.12 Acquisition of the furin cleavage site might be viewed as a ‘gain of function’ that enabled a bat CoV to jump into humans and begin its current epidemic spread. As we can see, the SARS-CoV-2 has evolutionary mechanisms that make it very infectious, but not so lethal to the point that it self-destructs with the death of the host it invades or attracts the attention of our immune system. Excessive lethality of an infectious agent impedes it from spreading from host to host, in cases such as MERS (Middle East respiratory syndrome) and Ebola, which despite extremely high lethality rates did not lead to pandemics like COVID-19 (Fig. 3).
Spike is subdivided into two subunits, S1 and S2, each with a different function. Source: Andersen et al.13.
The RBD of the spike in SARS-CoV-2 has a high affinity with ACE2 but is less accessible to the receptor because of the shaping of the trimers that comprise the structure of the spike. The dynamic state of RBD in coronavirus spikes explains this paradox; the RBD in coronaviruses can either be “up,” which allows it to connect with the receptor, or “down,” when it cannot bind with receptors. In the SARS-CoV-1 spike the RBD is usually “up” but in the spike protein in SARS-CoV-2 the RBD is generally “down”. For this reason, even though the RBD of SARS-CoV-2 has just as much affinity with ACE2 as the RBD in SARS-CoV-1, it is less accessible than in SARS-CoV-1,13 and as a result, SARS-CoV-2 is less exposed to human immune response. SARS-CoV-2 makes up for this weakness with at least three evolutionary advantages.
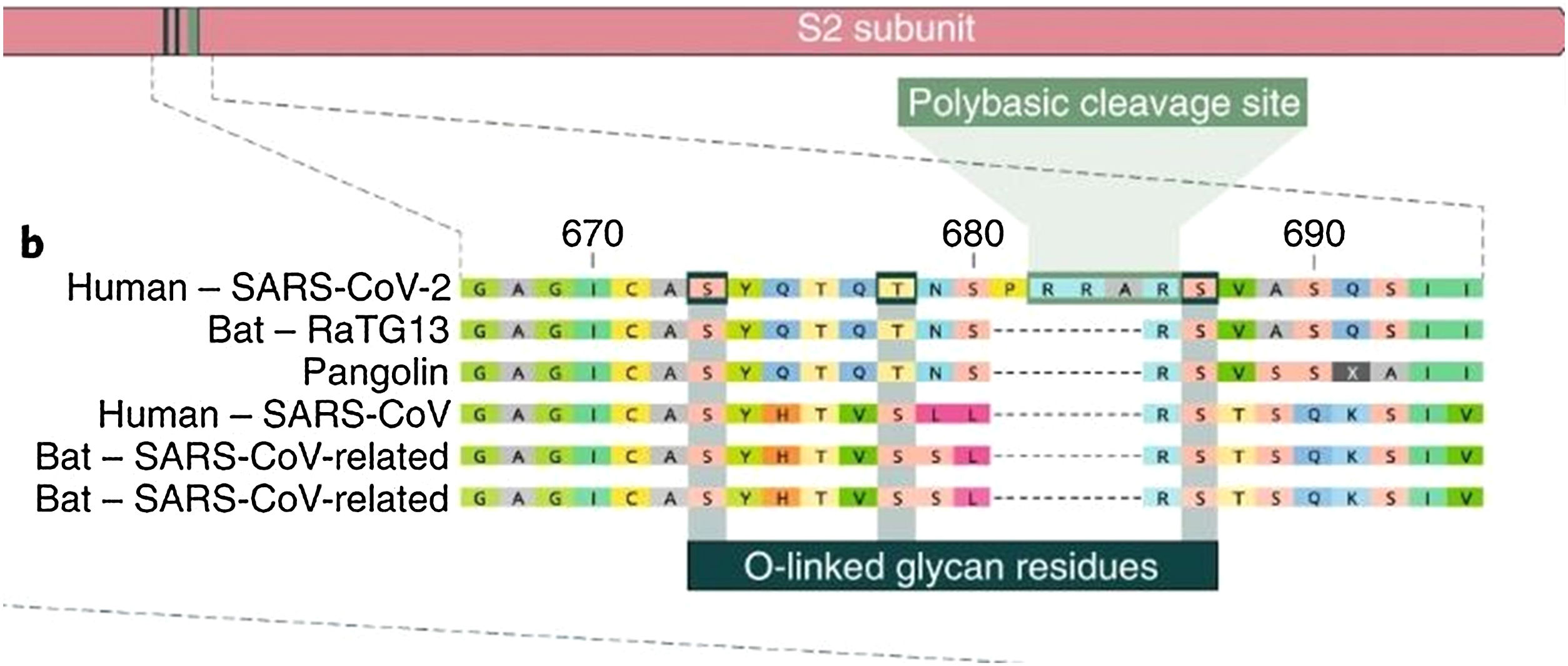
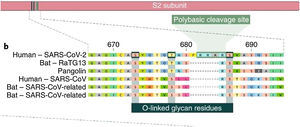
Of all the coronaviruses, only SARS-CoV-2 has a genetic sequence featuring an insertion of 12 base pairs (4 amino acids) in the spike, called the polybasic site. The four amino acids inserted into the sequence form an exposed loop, increasing the susceptibility of the S protein to cleavage mediated by protease, which facilitates infection by SARS-CoV-2. The insertion sequence also generates a cleavage site for the protease furin. The insertion sequence is unique and has not been found in any other known coronavirus, not even in the RaTG12 coronavirus (in bats), which is highly homologous to SARS-CoV-2. This insertion probably makes up for the poor accessibility of the SARS-CoV-2 RBD with ACE2.14
Pre-activation by the host proteasesPre-activation of the spike in SARS-CoV-2 by furin expands the virus’s ability to enter the types of cell lines that express ACE2, including pulmonary fibroblasts and epithelium. TMPRSS2 and lysosomal cathepsin activate the entry of SARS-CoV-2 and both have cumulative effects with the protease furin when SARS-CoV-2 enters the cell; in comparison, the entry of the SARS-CoV-1 virus is activated by TMPRSS2 and cathepsins, but not by furin.15
The presence of O-glycosidic bonds alongside the cleavage site
The insertion of a proline at the junction between S1 and S2 before the 12-base-pair insertion site results in the addition of O-glycosidic bonds in serines and threonines (S673, T678, and S686) flanking the cleavage site, which are unique to SARS-CoV-2. The O-glycosidic bonds can create a "mucin-like domain" that protects epitopes or key residues in the S protein of SARS-CoV-2. Various viruses use this domain as glycan shields for immunoevasion16 (Fig. 4).
Among all of the coronaviruses, only SARS-CoV-2 has the insertion of 4 amino acids in the spike located between s1 and s2, before the first s1/s2 cleavage site. Source: Andersen et al.13.
This question can be answered by comparing different virus genomes in order to find the nearest genetic homology. The SARS-CoV-2 shares 79.5% of its genome with SARS-CoV-1 and exhibits a remarkable 93.1% homology with the sequence of the RaTG12 virus isolated from a bat (Rhinolophus affinis) from Yunnan province, China, 2,000 km from Wuhan. Although the genome of RaTG13 has 96% homology with the genome of the SARS-CoV-2, its S protein differs in the RBD, indicating that it would not be able to effectively connect to human ACE2. Furthermore, direct contact between bats and humans is rare, so it is more likely that the transmission of SARS-CoV-2 to humans occurs through an intermediary host (and not directly from bats), as was the case in SARS-CoV-1 and MERS-CoV. As far as it is known, after Bat-CoV-RaTG13, the virus with the greatest genetic homology to the SARS-CoV-2 is the coronavirus that infects certain Malayan pangolins (Manis javanica).17 Considering only the homology of the RBD in SARS-CoV-2, the pangolin coronavirus genome sequence is more homologous than the bat coronavirus RaTG12. Six amino acids in the RBD have been described as determinants for an efficient connection between SARS-CoV-2 and ACE2, and the genome of the Malayan pangolin contains precisely these six amino acids in the same positions. The bat coronavirus RaTG13 has the same amino acid as the SARS-CoV-2 in only one of these six positions in the RBD, although the bat coronavirus RaTG13 is more homologous to the SARS-CoV-2 considering the genome as a whole. A new bat coronavirus identified in 227 bats collected from Yunnan province, China called RmYN02 is notable for the insertion of nitrogenous base pairs in the spike, which is very similar to the unique insertion of SARS-CoV-2 (the polybasic site). RmYN02 has a nucleotide sequence homology of 93.3% to the SARS-CoV-2, and this rises to 97.2% in the ORF1ab sequence, which is the most homologous to SARS-CoV-2. But in the RBD RmYN02 is less homologous (61.3%) to the SARS-CoV-2 and may not be able to connect to the ACE2. The bat coronavirus RATG13 may have obtained the RBD sequence that connects to the human ACE2 receptor through recombination with the coronavirus from the Malayan pangolin, and the polybasic site through recombination with the bat coronavirus RmYN02.18
This complex evolutionary pattern raises the hypothesis of recombination between multiple viral genomes impacted by natural selection before or during the evolutionary leap to human infection.
Could SARS-CoV-2 have been created in a laboratory?Various pieces of scientific evidence indicate that it is very unlikely that SARS-CoV-2 was created in a laboratory, either intentionally or by accident.
- 1
The genome of the SARS-CoV-2 has several differences from other coronaviruses besides the 12-base pairs for insertion. The virus with the greatest genetic homology is the bat coronavirus RaTG13, which shares "only" 96% of its genome with the SARS-CoV-2 (1200 different nitrogenous base pairs); no laboratory technology can simultaneously handle 1200 nitrogenous base pairs modifications.
- 2
The presence of unique sugar-fixing sites (O-glycosidic bonds) in the SARS-CoV-2 genome is another clue that the virus is natural. The sugars create a "mucin shield" that protects the virus from attack by the immune system. Since tissue culture plates in laboratories do not have an immune system, it is unlikely that such an adaptation would appear in a laboratory-grown virus, essentially toppling the hypothesis that the virus was multiplied in tissue culture.
- 3
The presence of an RBD very similar to that of SARS-CoV-2 in Malayan pangolins allows us to infer that this probably also occurred in the virus that was transmitted to humans, indicating that the polybasic insertion at the cleavage site may have occurred during human-to-human transmission.
- 4
The RBD in the SARS-CoV-2 is different from that of the SARS-CoV-1, and the SARS-CoV-2’s connection to ACE2 is not ideal, which implies that other connection mechanisms (“down” RBD and the polybasic cleavage site providing pre-activation via furin) resulted from natural selection. For this reason, not only the understanding that its “strengths” evolved naturally but also the existence of “weak points” in the SARS-CoV-2 suggest that the virus was not artificially manipulated.
The coronavirus genome is one of the largest among the RNA viruses, and it is natural for variations to occur. However, researchers have found that the coronavirus is mutating relatively slowly compared to other RNA viruses. This is partly because proofreading proteins like ExoN (encoded by the non-structural protein Nsp14) are able to correct some errors. The average mutation rate is approximately 8 × 10−4 substitutions per nucleotide per year. Since the RNA of the SARS-CoV-2 has almost 30,000 nucleotides, approximately 23,127 substitutions/year are expected, roughly 2 per month.19 This data was recently obtained for the strains present in the Brazilian population, estimated at 1.3 × 10−3 substitutions per nucleotide per year, the equivalent to an average of 33 variations per year, which is slightly higher than the global average. Even so, the mutation rate is lower than in SARS-CoV-1 or influenza. This does not mean that new strains of SARS-CoV-2 even more pathogenic than those currently circulating may not emerge. One reason (but not the only one, and not a sufficient criterion) to suspect that a certain mutation is being selected because it has an important phenotypic consequence is to monitor whether it is attaining a high frequency in the population. Thanks to the extraordinary worldwide collaborative efforts to sequence and publicly share information on the thousands of SARS-CoV-2 genomes,20 fourteen variants with slightly higher frequencies have already been identified.21 The majority of these variants have no functional effect, follow a neutral evolutionary dynamic, and often involve mutational hotspots. The most notable among those variants that have reached a relevant frequency is D614G, in which guanine is replaced with adenine in position 23,403 at terminal C of S1 in the spike. In Europe, where G614 began its expansion, the D614 and G614 forms were co-circulating at the beginning of the epidemic, although D614 was more common in most of the countries surveyed. In March 2020, G614 became increasingly common throughout Europe, and by April it dominated the sampling at that time. There is growing evidence that this variant may have an effect on transmission capacity and even on the severity of the COVID-19 infection. The G614 mutation may decrease interaction between the S1 and S2 units, facilitating S1’s detachment from S2 (connected to the viral membrane). Some studies suggest that G614 spread so rapidly through the region because the SARS-CoV-2 with this variant would be more infectious, and perhaps even associated with a higher viral load, but these same studies found no difference in hospitalization rates.22 Other authors suggest that the predominance of this variant as the pandemic spread (and its low prevalence during the initial stages) could be related to the deletion of a single nucleotide in the TMPRSS2 gene; this deletion is frequent in Europeans and North Americans, but rare in Asia. The G614 variant introduces a cleavage site for the protease TMPRSS2. The presence of this variant of a single nucleotide in the TMPRSS2 gene could interfere in the virus-host relationship.23 It is still not clear whether the G614 variant is a result of natural selection or what it may really mean for the COVID-19 pandemic. Will it make the pandemic more difficult to control? Is it making the SARS-CoV-2 more infectious? Will it affect the development of therapies and treatments? Further studies are needed to address these questions.24
Molecular epidemiology of the SARS-CoV-2 in BrazilFrom the start of the pandemic, Brazil has stood out for its ability to quickly sequence the SARS-CoV-2 genome in Brazilians. These first efforts, since they took place at the very beginning of the pandemic and involved small samples, mirrored another form of propagation through transmission coming from the outside. At first, works sequencing SARS-CoV-2 genomes in the Brazilian population were published, reflecting the reality of patients in São Paulo,5 Minas Gerais,25 and Amazonas,26 for example. More recently a study was published with a larger sample including patients from various regions of Brazil and reflecting local community transmission. To investigate the strains of SARS-CoV-2 circulating in the country, researchers analyzed the genomes of SARS-CoV-2 collected from Brazilian patients infected during the first two months of the COVID-19 outbreak. The samples were collected between February 29 and April 28, 2020, from individuals living in 10 Brazilian states in the southeast (Rio de Janeiro and Espírito Santo), midwest (Federal District), north (Acre, Amapá, and Pará), northeast (Alagoas, Bahia, and Maranhão) and south (Santa Catarina). Only seven individuals reported international travel or contact with people who had traveled. Six different strains of SARS-CoV-2 (A.2, B.1, B.1.1, B.2.1, B.2.2, and B.6) were detected in the samples. The majority of these Brazilian sequences of SARS-CoV-2 were classified as the B.1 clade (95%, n = 90), particularly the B.1.1 subclade (92%, n = 87). The prevalence of the B.1.1 subclade in this sample (92%) was much higher than that observed in other earlier Brazilian sequences (36%). The B.1.1 clade was the only lineage detected in individuals without recent international travel, while four different strains (B.1, B.1.1, B2.1, and B.6) were detected among the six individuals who reported recent international travel (imported cases) and their contacts. Besides sharing the three nucleotide mutations characteristic of the B.1.1 clade (G28881A, G28882A, G28883C), the sequences in the B.1.1.EU/BR and B.1.1.BR clusters harbor a non-synonymous T29148C mutation in the nucleocapsid protein (I292 T); another non-synonymous T27299C mutation in ORF6 (R33 T) was shared only by sequences in the B.1.1.BR lineage. The T29148C and T27299C mutations were not detected in the other 7551 B.1.1 genomes found worldwide, corroborating the hypothesis that they are synapomorphic traits of the B.1.1.EU/BR and B.1.1.BR clades, respectively. The B.1.1.EU/BR clade was highly prevalent in Minas Gerais and was also detected in the Federal District, while the B.1.1.BR clade was predominant in Rio de Janeiro and also identified in some samples from the north, midwest, and northeast regions. Notably, none of these strains were detected in the most populous state, São Paulo.27
Another important genomic surveillance study was performed by Brazilian researchers and recently published in the journal Science, exhibiting broad knowledge about the epidemic transmission and evolutionary trajectory of the SARS-CoV-2 lineages in Brazil. These authors sequenced 427 genomes of patients residing in 85 municipalities of 18 states across all regions of Brazil and identified over 100 international introductions of strains into Brazil; São Paulo accounted for 36% of these imports, Minas Gerais 24%, Ceará 10%, and Rio de Janeiro 8%. They estimated that 76 of the Brazilian strains of SARS-CoV-2 fall into three clades and were introduced from Europe between February 22 and March 11, 2020. This group also showed that clade 1 circulates primarily in the state of São Paulo, clade 2 in 16 states, and clade 3 in the state of Ceará. Clades 1 and 2 represent the majority of these lineages in the states of Pará and Amazonas; the latter group is associated with multiple entries, both from within Brazil as well as international. They concluded that changes in mobility may impact the global and the local transmission of SARS-CoV-2, in a sophisticated and elegant combination of genomic and mobility data allied with traditional epidemiological surveillance strategies.28
These studies clearly demonstrate the significant benefits to the country from investment in research, particularly in terms of aspects that might not attract the attention of researchers abroad. Understanding the molecular epidemiology of the SARS-CoV-2 in a country that spans an entire continent, like Brazil, has a major impact on understanding its evolution and developing public health strategies to combat the virus.
What is the role of the human genome in COVID-19?All infections have consequences that are related not only to the infectious agent but also to the host. COVID-19 has been shown to be extremely heterogeneous and consequently challenging with regard to understanding its multiple clinical and epidemiological aspects. It is no exaggeration to say that seven months after the first case detected in the world we have more questions than answers.
- ✓
Why children are infected less frequently and less severely?
- ✓
Why do some healthy young adults get seriously ill?
- ✓
Why do many people remain asymptomatic?
- ✓
Why do many people develop mild cases while others are so serious?
- ✓
Why do more men die of COVID-19 than women?
- ✓
Why are certain ethnic groups more affected than others?
- ✓
Why is such a large portion of infections associated with only a few individuals?
- ✓
Why do some patients remain RT-PCR positive?
- ✓
Why do some take so long to become IgG positive?
- ✓
Why do some respond better to certain treatments?
- ✓
Why do some take so long to recover from the disease?
These are just some of the questions that the scientific community still does not know how to answer. At least some of the answers to these questions are expected to come from studies that investigate not only the genome of the virus but also the human genome and its interactions with the SARS-CoV-2 genome. Several genes are natural candidates for study, including those that are known to play a crucial role in our immune defense system (as in the case of MHC/HLA and CCR5), those that encode for the membrane receptors that bind to the spike in SARS-CoV-2 (like ACE2), and those that encode for proteases that activate SARS-CoV-2 (like TMPRSS2 and lysosomal cathepsins). To this end, an international research consortium on genetic factors in the human genome has been organized.29 The most relevant work conducted so far has used techniques that investigate millions of variants in the human genome, in patients with COVID-19 and controls, without indicating candidate genes a priori.30 This study assessed 1980 patients with severe COVID-19 (defined as hospitalization for respiratory insufficiency and confirmed RT-PCR for SARS-CoV-2) and 2381 participants as controls from Italy and Spain. Greater risk was found among persons with blood type A than other blood types (odds ratio: 1.45; 95% CI, 1.20–1.75; p = 1.48 × 10−4), and O type blood had a protective effect compared with other types (odds ratio: 0.65; 95% CI, 0.53–0.79; p = 1.06 × 10−5). On chromosome 3, more specifically at 3q21.31, the association peak spanned a cluster of six genes (SLC6A20, LZTFL1, CCR9, FYCO1, CXCR6, and XCR1), several of which have functions that may be relevant to COVID-19.29 More detailed studies are needed to investigate whether these findings can be replicated and also to understand how and why these genes could impact the relationship between the genome of human host and SARS-CoV-2. Even more intriguing, this cluster of six genes in 3p21.31 was shown to be derived from part of the Neanderthal genome that was inherited by modern humans.31 Only future studies will determine the evolutionary significance of this finding.
One of the more surprising and intriguing clinical/epidemiological aspects of COVID-19 is the indisputable fact that children rarely experience the more severe forms of the disease, but some display a unique Multisystem Inflammatory Syndrome. These aspects have recently been discussed in the Editorial section of this journal.32 Emergent studies that indicate substantial differences in gene expression of the ACE2 membrane receptor and TMPRSS2 (in nasal as well as lung epithelial cells) are particularly important in pediatrics. Children have lower expression of these proteins, which are the real entryways to the body for SARS-CoV-2, and that may partly explain why COVID-19 evidently spares the pediatric population compared to adults and the elderly in particular.33,34
Not only genetic variants but also interactions between SARS-CoV-2 and human proteins are being investigated, especially in the cells of the heart and lungs, the two organs most affected by COVID-19. Studies of the “cardiac interactome” indicate the importance of microRNA as biomarkers that predict severity,35 and research on the interactome of lung cells showed potential ACE2 regulators in the human lung, including genes related to modifications of histones such as HAT1, HDAC2, and KDM5B.36
Genetic vaccinesOver 320 vaccines for COVID-19 are being developed, and 32 of these are already in clinical trials in humans. The great novelties are “genetic” vaccines, which thanks to genetic engineering and vectors carry copies of SARS-CoV-2 genes (particularly, for the spike) or those that directly inject synthetic mRNA from SARS-CoV-2 to evoke an immune response. These mRNA vaccines lower the risk of integrating into the host’s genome, an advantage of mRNA over DNA vaccines.37 These genetic vaccines for COVID-19 were developed at an impressive speed, since from previous Coronavirus research, the antigen role of the spike protein and antibodies against it were already known to be fundamental in immunity. A contribution came from the record-breaking speed with which the SARS-CoV-2 genome was sequenced after confirmation of the first cases in China, in turn leading to significant progress in the nucleic acid vaccine platforms that allow the quick production of thousands of copies of the vaccine. One of these mRNA vaccines was the first to enter clinical trials and began to be developed as soon as the genetic sequence of SARS-CoV-2 was published on January 10, 2020. The first 45 volunteers received the vaccine in phase 1 trials on March 16, 2020, 66 days after the publication of the genomic sequence. The phase 1 results of this vaccine were published on July 14, 2020.38 Even before the phase 1 results publication, phase 2 trials began on May 11, 2020, involving 300 young adults and 50 adults over the age of 55. Phase 3 trials that will involve 30,000 people are underway at the time of writing, and 23,947 participants have enrolled as of Friday, September 11, 2020. The time needed to test and produce vaccines (usually around a decade) was reduced to one year.
ConclusionsUnprecedented global efforts to sequence the SARS-CoV-2 genome in record time and study it in detail could make all the difference in how we deal with this threatening pandemic. Investments in local industrial installations that can produce reagents for molecular biology and pharmaceuticals, as well as in genetic research and in human and institutional resources for studying genomes have clearly become a strategic objective for every country that wants to be self-sufficient now and during future pandemics. Independence in this field of knowledge is not a matter of status or elitism, but rather is vital for understanding the epidemiology and spread of these infectious agents among our population, quickly recognizing potential evolutionary changes, diagnosing individuals through the genomic testing of these infectious agents, understanding clinical and therapeutic variability, and developing safe and effective vaccines with the degree of urgency that is clearly necessary to combat COVID-19. The main limitations of this review come from what is still unknown regarding the genetics of COVID-19, the unprecedented volume and speed of scientific information about COVID-19 with new data accumulating on a daily basis, and the fact that we are writing it in the middle of the pandemic. For sure there will be valuable new information not included in this review by the time it is published. Even so, this narrative review should be useful as a basis for understanding the genetics of COVID-19.
FundingThe author declares that he has not received any funding or research grants in the course of the study, research, or assembly of the manuscript.
This manuscript has not been published previously and is not being evaluated for publication in any other journal. Publication was approved by all authors and tacitly or explicitly by the responsible authorities where the study was conducted. If this manuscript is accepted, it will not be published elsewhere in this same format, in English or any other language, including electronically, without prior written consent from the copyright holder. The originality of this manuscript can be verified using the CrossCheck plagiarism detection service.
He hereby confirms the availability of the data supporting this study when submitting this article.
Conflicts of interestThe author declares no conflicts of interest.