The aim of this study was to evaluate the QuickVue® RSV Test Kit (QUIDEL Corp, CA, USA) as a screening tool for respiratory syncytial virus in children with acute respiratory disease in comparison with the indirect immunofluorescence assay as gold standard. In Brazil, rapid antigen detection tests for respiratory syncytial virus are not routinely utilized as a diagnostic tool, except for the diagnosis of dengue and influenza.
MethodsThe authors retrospectively analyzed 486 nasopharyngeal aspirate samples from children under age 5 with acute respiratory infection, between December 2013 and August 2014, the samples were analyzed by indirect immunofluorescence assay and QuickVue® RSV Test kit. Samples with discordant results were analyzed by real time PCR and nucleotide sequencing.
ResultsFrom 313 positive samples by immunofluorescence assays, 282 (90%) were also positive by the rapid antigen detection test, two were positive only by rapid antigen detection test, 33 were positive only by immunofluorescence assays, and 171 were positive by both methods. The 35 samples with discordant results were analyzed by real time PCR; the two samples positive only by rapid antigen detection test and the five positive only by immunofluorescence assays were also positive by real time PCR. There was no relation between the negativity by QuickVue® RSV Test and viral load or specific strain. The QuickVue® RSV Test showed sensitivity of 90%, specificity of 98.8%, predictive positive value of 99.3%, and negative predictive value of 94.6%, with accuracy of 93.2% and agreement κ index of 0.85 in comparison to immunofluorescence assay.
ConclusionsThis study demonstrated that the QuickVue® RSV Test Kit can be effective in early detection of Respiratory syncytial virus in nasopharyngeal aspirate and is reliable for use as a diagnostic tool in pediatrics.
Avaliar o teste QuickVue® RSV Test Kit (QUIDEL Corp, CA, EUA) para o diagnóstico rápido do vírus sincicial respiratório em crianças com doença respiratória aguda, comparando-o com a imunofluorescência indireta como padrão ouro. Visto que, no Brasil, testes rápidos para detecção de antígenos para vírus sincicial respiratório não são rotineiramente utilizados como ferramenta de diagnóstico, exceto para Dengue e Influenza.
MétodosUm total de 486 amostras de aspirado de nasofaringe de crianças menores de 5 anos com doença respiratória aguda, coletadas entre dezembro de 2013 e agosto de 2014, foram analisadas por imunofluorescência e pelo teste QuickVue®. Amostras com resultados discordantes entre os métodos foram submetidas a PCR em tempo real e sequenciamento.
ResultadosDas 313 amostras positivas por IFI, 282 foram positivas no teste rápido (90%), 2 amostras foram positivas apenas no teste rápido (0.6%), 33 apenas na imunofluorescência (10.5%) e 171 foram negativas em ambos os métodos. As 35 amostras com resultados discordantes foram testadas por PCR em tempo real, sendo que duas que foram positivas apenas no teste rápido e 5 apenas na imunofluorescência confirmaram-se positivas. Não houve relação entre a ausência de positividade no teste QuickVue® com a carga ou com a cepa viral. O teste QuickVue® mostrou sensibilidade de 90.1%, especificidade 98.9%, valor preditivo positivo 99.3%, valor preditivo negativo de 94.6%, acurácia de 93.2% e índice κ de concordância de 0.85 em comparação à imunofluorescência.
ConclusõesNosso estudo demonstrou que o teste QuickVue® RSV pode ser efetivo na detecção precoce do vírus sincicial respiratório em amostras de aspirado de nasofaringe e é confiável como uma ferramenta de diagnósticos em pediatria.
Respiratory syncytial virus (RSV) is known as a major infectious agent of respiratory tract infections in children worldwide.1 Most children are infected in the first year of life, but virtually all will be exposed by age 2.2 Reinfections are common throughout life, depending on the level of neutralizing antibodies in the serum, but complications in lower respiratory tract infections are more common in primary infection.3 RSV is primarily replicated in superficial portions of the respiratory tract, until it spreads through the epithelium, forming a syncytia-like cytopathic effect. RSV has two known sub-groups: A and B, and it can cause a range of clinical conditions, from the common cold to bronchiolitis and pneumonia, which are caused by necrosis of the bronchi and bronchioles. The World Health Organization (WHO) estimates that RSV infects 64 million people and causes 160,000 deaths per year worldwide.4 The seasonality of the virus varies, and it is often detected throughout the year. However, it is known that the highest incidence occurs in the winter season.
The viral diagnosis of RSV can be accomplished by a number of methods, including: cell culture, immunofluorescence assays (IFA), immune chromatographic assays (rapid antigen detection tests: RADTs), and polymerase chain reaction (PCR), including conventional and real-time PCR assays. In the last decade, the molecular methods have been used as the gold standard, because of their specificity and ability to simultaneously detect different viruses.5 In Brazil, although there are at least four RADTs available for RSV detection (BD-Directigen™EZ-RSV [Becton, Dickinson and Company®, NJ, USA]; SASTM RSValert [Medivax®, RJ, Brazil]; Alere™BinaxNow®RSV [Alere™, USA]; and QuickVue® RSV Test Kit [QUIDEL Corp., CA, USA]), they are not routinely utilized in the country as a reliable diagnostic for viral infections, such as RSV, with the exception of human immunodeficiency virus (HIV) and dengue tests,6,7 which are widely used.
The rapid test to be evaluated in this study (QuickVue® RSV Test Kit, Quidel®)provides a result in 15min, compared to approximately 90min for a conventional IFA test and 2–3h for enzyme (ELISA).8 Quickly identifying the etiologic agent of respiratory diseases, such as bronchitis and bronchiolitis, is an important step toward reducing the use of antimicrobials, especially in hospitalized children,9 and can also indicate the most appropriate method of isolation and treatment.
The QuickVue® RSV test is a dipstick immunoassay, which allows for the rapid, qualitative detection of RSV antigen (viral fusion protein) directly from nasopharyngeal swab, nasopharyngeal aspirate, or nasal/nasopharyngeal wash specimens from symptomatic pediatric patients. The test is intended for use as an aid in the diagnosis of acute respiratory syncytial viral infections.
The aim of this study is to evaluate sensitivity, specificity, and importance of the QuickVue® RSV Test Kit as a RADT for screening and improvement of RSV diagnosis in the nasopharyngeal aspirate of children with acute respiratory disease compared with indirect IFA, for use in hospitals and pediatric clinical practices.
Materials and methodsThe authors retrospectively analyzed nasopharyngeal aspirate (NPA) samples from 486 children – 274 boys (56.4%) and 212 girls (43.6%) – under five years old with symptoms of acute respiratory infection (ARI), including upper and lower respiratory tract disease, in the period between December 2013 and August 2014, who had ARI and were cared for in the emergency room, as outpatients, or hospitalized in the Pediatrics Department, University Hospital, University of São Paulo (HU-USP).
Clinical samples were collected during the RSV outbreak of 2014 using mucus trap. Immediately following collection, the samples were sent to the University of São Paulo Hospital Laboratory and screened by indirect IFA using a commercial Kit Light Diagnostics™ Respiratory Panel Viral Screening and Identification (IFA Chemicon®, Merk Millipore Corp., USA) following the manufacturer's instructions. These samples were then sent to the Clinical and Molecular Virology Laboratory at the Institute of Biomedical Sciences of the University of São Paulo, where they were frozen by cryogenics, and stored at −80°C until the time of testing. All the samples were registered at the Virology Laboratory Biorepository, and all ethics guidelines for human experimentation were strictly observed and approved by the Ethics Committee on Research with Human Beings at the University of São Paulo (USP).
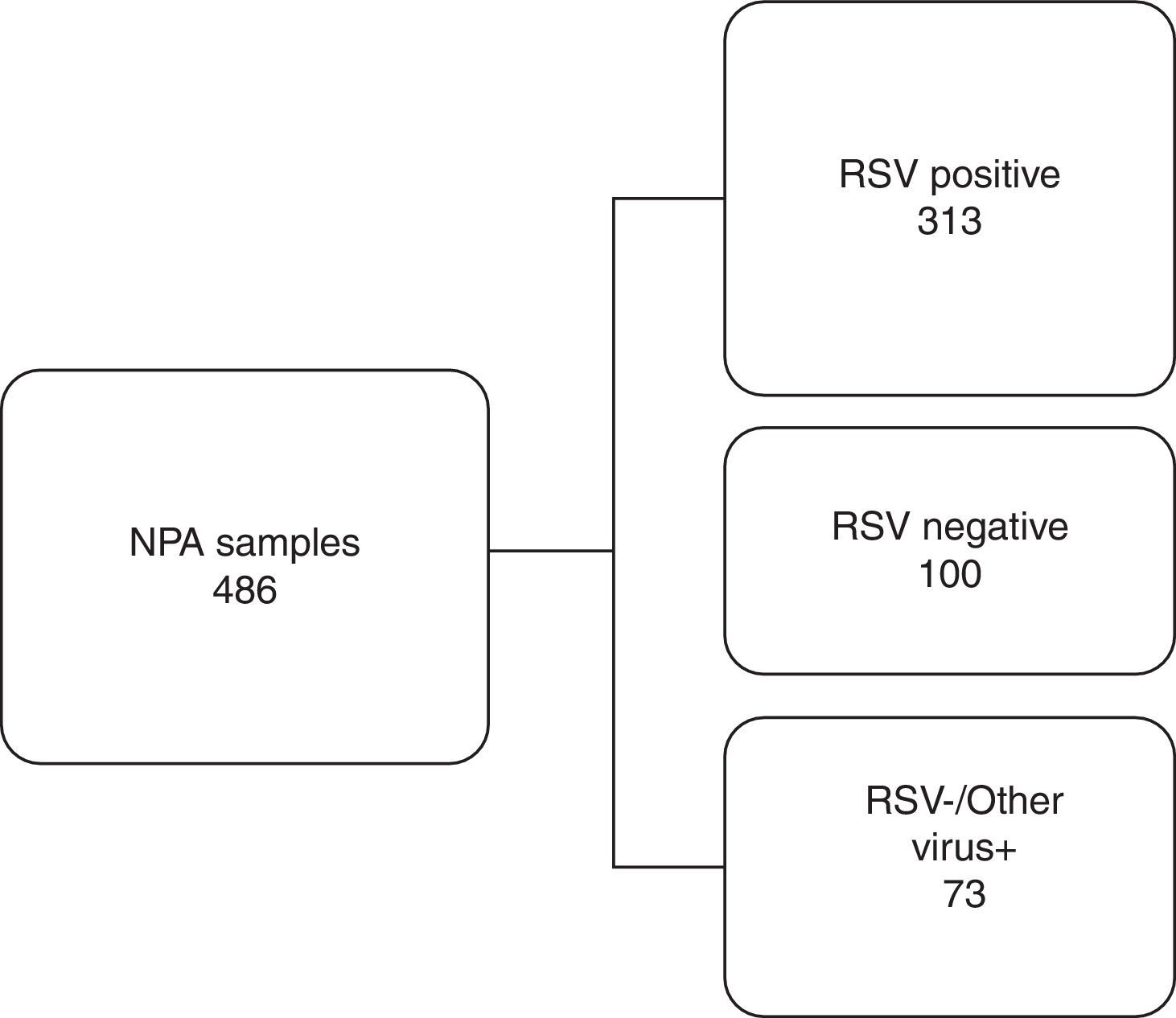
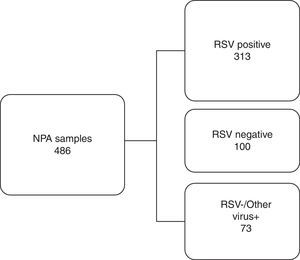
Based on the IFA results, 486 NPA samples distributed during all the months of the season were selected, corresponding to 50.4% of all the samples analyzed. They were classified into three distinct groups as shown in Fig. 1. To reduce or eliminate any bias, the QuickVue® RSV assay was performed and analyzed in a blind trial, and results were compared with the original results from the IFA Kit.
Work flowchart. Nasopharyngeal aspirate samples were divided into three distinct groups screened by immunofluorescence assay and compared with rapid antigen detection test for respiratory syncytial virus. RSV, respiratory syncytial virus; NPA, nasopharyngeal aspirate; +, positive; −, negative.
In order to confirm the results, samples with discrepant results were subsequently tested by real-time quantitative reverse transcription PCR (F protein region) and by traditional PCR (G and F protein regions).
Samples were extracted on the NUCLISENS® easyMag® platform (bioMérieux®, MA, USA) and real-time PCR performed on ABI 7300 instrumentation utilizing the AgPath-ID One-Step RT-PCR master mix kit (Ambion®, TX, USA) following the manufacturer's instructions.
For traditional PCR, the authors used a reaction mixture containing 10μL of cDNA, 5μL of PCR buffer reaction 10× concentration (50mM Tris–HCl [pH 9.0], 1.0μL of MgCl2 (50mM KCl, 20mM [NH4)2SO4]), 2.5mM of each dNTPs, 10pmol of each primer for RSV (Table 1), 0.6U of Taq DNA Polymerase (platinum taq DNA polymerase, Invitrogen, CA, USA) and water, resulting in a final volume of 50μL. Amplification of protein G and F was performed in a GeneAmp PCR System 9700 (Applied Biosystems, Inc., CA, EUA) thermocycler according to the following program: 95°C for five min, followed by 40 cycles of 95°C for 30s, 54°C for 30s, 72°C for 90s, and one last seven min cycle at 72°C. Approximately 1894bp products of G gene10–12 and 1685bp products of F gene13 were purified with a commercial kit (Exosap, GE®Tech, CA, USA), according to the manufacturer's instructions. Sequence reactions of the G gene10–13 and the F gene13,14 were subjected to electrophoretic separation for primary data collection in a 3130 DNA sequencer (Applied Biosystems, Inc., CA, EUA), using a fluorescent dye terminator kit (Applied Biosystems, Inc.). Sequences obtained were edited with the Sequence Navigator program version 1.0 (Applied Biosystems, Inc., CA, EUA) and aligned using the program Megalign (Lacergene, DNA STAR, Inc., WI, USA). A maximum likelihood phylogeny of G protein was estimated using MEGA 615,16 with gamma distribution (TN93+G) for the RSV_A data set and model with gamma distribution (HKY+G) for HRSVB.17 The phylogenetic tree that resulted was mid-point rooted.
Primers used for traditional polymerase chain reaction (PCR) and sequencing of G and F respiratory syncytial virus (RSV) proteins.
| Primer name | Target protein | Genome position | Reaction | Sequence 5′-3′ | Reference |
|---|---|---|---|---|---|
| FV-a_AB | G | 186–163/Gene F | PCR | GTTATGACACTGGTATACCAACC | Zheng et al.10 |
| SH+_AB | G | 3924–3948/Gene M | PCR | CAGCTACACGTTTTTCAATCAAACC | Lima et al.11 |
| OG1+_21a | G | 1–21/Gene G | SANGER | GGGGCAAATGCAACCATGTCC | Trento et al.12 |
| GAB+a,b | G/F | 504–524/Gene G | PCR | YCAYTTTGAAGTGTTCAACTT | Peret et al.13 |
| BO2–_ABb | F | 1216–1199/Gene F | PCR | ATCTGTTTTTGAAGTCAT | Arbiza J.c |
| F1AB+b | F | 3–22/Gene F | SANGER | GGTTGGTATACCAGTGTCATAAC | Modif. Peret et al.14 |
| FP2+_ABb | F | 573–595/Gene F | SANGER | TTAACCAGCAAAGTGTTAGACC | In House |
G, glycoprotein; F, fusion protein; M, matrix protein.
The criteria used for the performance assessment of QuickVue® RSV assay were sensitivity, specificity, positive and negative predictive values, and positive/negative diagnostic likelihood ratios (DLRs).18,19 IFA was considered as the gold standard for these analyses. Samples positive by IFA were considered to be true positives, and samples that were negative by IFA were considered to be true negatives. For accurate evaluation of the agreement between the methods, the kappa index (κ) was also assessed.
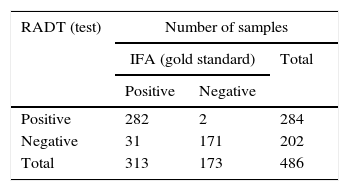
ResultsRADTOf the 313 NPA samples positive for RSV by IFA, 282 (90%) were also positive by the rapid test (QuickVue® RSV test), thus the RADT test revealed 31 false negative results. All the 100 NPA samples negative for RSV by IFA were also negative by the rapid test (100%). Of 73 NPA samples that were negative for RSV but positive for other respiratory virus (AdV or PIV 1, 2, 3 or Flu A and Flu B) by IFA, 71 (97.26%) were confirmed by the rapid test, and two (2.74%) had divergent results, revealing two false positive results (Table 2A).
Results of respiratory syncytial virus (RSV) positive and negative by indirect immunofluorescence assay (IFA) and rapid antigen detection test (RADT) and the totality of divergent samples tested in this study. (A) Rapid-antigen detection test results compared with IFA. (B) Discrepant results. (C) Comparison of rapid test evaluation parameters including sensitivity, specificity, predictive positive value (PPV), negative predictive value (NPV), and positive/negative diagnostic likelihood ratio (DLR), for QuickVue® RSV test compared to IFA, used as the gold standard.
| RADT (test) | Number of samples | ||
|---|---|---|---|
| IFA (gold standard) | Total | ||
| Positive | Negative | ||
| Positive | 282 | 2 | 284 |
| Negative | 31 | 171 | 202 |
| Total | 313 | 173 | 486 |
| Results | Number of samples |
|---|---|
| IFA+, RADT−, qPCR+ | 26 |
| IFA−, RADT+, qPCR+ | 2 |
| IFA+, RADT−, qPCR− | 5 |
| IFA−, RADT+, qPCR− | 0 |
| Parameter | 95% CI |
|---|---|
| Sensitivity (%) | 90.1 (86.8–93.4%) |
| Specificity (%) | 98.8 (97.2–100) |
| PPV (%) | 99.3 (98.3–100%) |
| NPV (%) | 94.6 (79.6–89.6) |
| Positive DLR | 77.9 (19.6–309.26) |
| Negative DLR | 0.1 (0.071–0.14) |
The 33 samples with discordant results between the two methods were analyzed by real-time PCR (Table 2B).
Twenty-six NPA samples that were previously considered positive by IFA and negative by rapid test were confirmed as positive for RSV by real-time PCR. The two NPA samples considered negative by IFA and positive by the rapid test were actually positive by qPCR. And the five NPA samples considered positive by IFA and negative by the rapid test were negative by qPCR. The real-time PCR threshold cycle (Ct) values of the 26 positive samples ranged from 14 to 37 cycles.
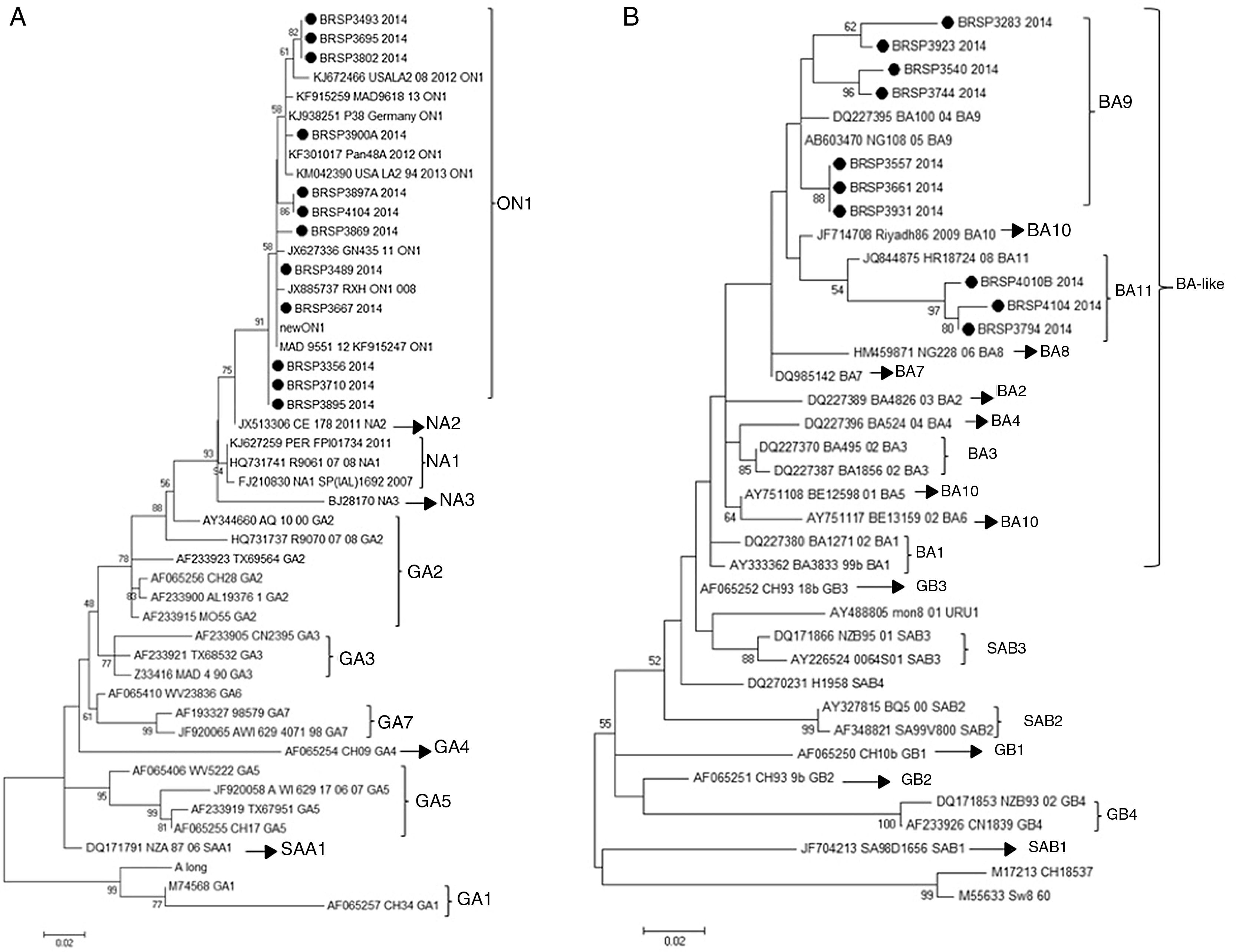
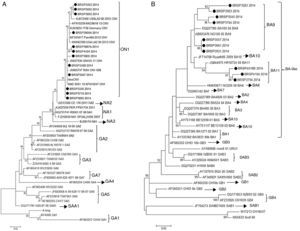
In order to understand if the differences between the tests were related to a specific genotype (RSV_A or RSV_B) or a genetic variability in the F protein of the virus, the G protein of all 26 samples with divergent results were sequenced and there were 22 consensuses for further analysis. After sequencing, the results showed that all the 12 RSV_A sequences were from the novel genotype ON1 and all ten RSV_B sequences were from the new BA11 (three) and BA9 (seven) genotypes (Fig. 2).
Human respiratory syncytial virus (HRSV) maximum likelihood phylogenetic tree based on nucleotide sequences of the G protein of the discrepant samples from this work, indicated as BRSP, and worldwide distributed strains obtained from GenBank. GenBank accession numbers are given in each taxon followed by the correspondent name of the strain. Initial tree(s) for the heuristic search were obtained by applying the neighbor-joining method to a matrix of pairwise distances estimated using the maximum composite likelihood (MCL) approach. The trees were drawn to scale, with branch lengths measured in the number of substitutions per site. Numbers at the internal nodes represent the bootstrap probabilities (5000 replicates). Only bootstrap values >50 are shown. (A) HRSVA. The evolutionary history was inferred by using the maximum likelihood method based on the Tamura–Nei model.15,16 The tree with the highest log likelihood (−1303.6711) is shown. (B) HRSVB. The evolutionary history was inferred by using the maximum likelihood method based on the Hasegawa–Kishino–Yano model.17 The tree with the highest log likelihood (−1207.4096) is shown. The analyses were conducted in MEGA 6.15
In order to verify if the negativity in RADT of positive samples with high load viral were associated to a specific mutation in the fusion (F) protein of RSV, the first 1200 nucleotides of the gene from the 26 samples with divergent results were sequenced and did not reveal a specific mutation that could be associated with the negativity of the samples in the RADTs (data not shown).
Statistical analysisThe performance of the QuickVue® RSV test compared to IFA are represented in Table 2. The accuracy was 93.2% (95% CI: 91.0–95.4%) and the agreement κ index between the methods was 0.85 (95% CI: 0.945–0.769), which was ranked as almost perfect,20 with p<0.001.
The analysis of the discordant and concordant results between the methodologies showed no significance (p=0.62, p=0.065 in the chi-squared test, respectively).
DiscussionThe aim of this study was to prove that the rapid test is efficient for RSV detection and to encourage its use in Brazil, as it is currently done for dengue, influenza, and HIV.
The hallmark of RSV infection is bronchiolitis,21 and its diagnosis is generally made by clinical condition.22 Although not recommended, the prescription of antibiotics for bronchiolitis is an extremely common practice in hospitalized children,9,23 and their use is associated with an increase in the cost and time of hospitalization24 and an increase of the risk of development of resistant bacterial strains.25 In severe cases, especially in intensive care units, antibiotics are commonly used to avoid a potential bacterial infection, although the risk in healthy infants is uncommon.26 A systematic review that analyzed four studies involving children attending the emergency room suggested that diagnostic tests could reduce the prescription of antibiotics,24 as was verified in a study with infants under 1 year of age hospitalized with bronchiolitis and tested positive for RSV by IFA that described a significant decrease in the use of antibiotics.27 If the diagnostic test is positive, it can contribute to prevent the use of antibiotics. In doubtful cases that are not severe, the patient should stay under observation.
Since the result using a rapid test is available in 15min and it is performed by the doctor without the need of laboratory equipment, the rapid test would be used for this screening, as a point of care assay.
In this context, the authors’ work showed that the QuickVue® RSV Test is reliable for this task, as can be seen by the results of the performance in comparison to indirect immunofluorescence assay (IFA) used as gold standard in this study. All the parameters analyzed were high (greater than 90% – sensitivity of 90%, specificity of 98.8%, PPV of 99.3%, and NPV of 94.6%) and were similar to those of other studies that compared RADT with IFA as the gold standard, which ranged from 73% to 93% sensitivity and 84% to 100% specificity.28 In contrast, in 2015 Leonardi et al.29 evaluated four RSV direct antigen detection assays and their results presented only 57.5% sensitivity for QuickVue®, although this difference may be due to the comparison with RT-qPCR, which has a higher sensitivity and specificity than IFA.
In addition, the overall agreement (kappa index) and accuracy between the two tests were considered good, and the negative and positive percent agreement (173/202 and 284/313 respectively) showed no significant difference (p>0.05). Only 33 samples showed divergent results between the tests; they were analyzed by RT-qPCR in order to confirm the results and to correlate the sensitivity of the test with a low viral load, as it was observed by Tuttle et al.30 Twenty-six samples continue to be unsolved after this analysis, and the authors also could not explain the differences by the load viral, since the qPCR threshold cycle ranged from 14 to 37. Maybe these false negative results could be related to the storage of frozen specimens before their analysis.
Next, in order to understand if these differences were related to a specific genotype (RSV_A or RSV_B) or a genetic variability in the F protein of the virus, the G and F genes were sequenced. The G sequences analysis showed that they belonged to genotypes ON1 and BA-like insertion (variants BA11 and BA9). However, other samples in this study that showed positive results by QuickVue® RSV Test were sequenced (data not shown) and belonged to the same genotypes, proving that the negativity is not related to these genotypes. This study did not identify any mutation in the F gene that could be responsible for the results. The final approach was to look for inhibition caused by chemical elements in the samples as an inhibition, nonetheless false negative specimens were diluted (1:5 and 1:10), but remained negative.
In conclusion, the QuickVue® RSV Test Kit displayed high predictive values and likelihood ratios, and it proved to be effective in early detection of RSV. Nevertheless, depending on the patient's improvement, it is recommended that the test must be repeated in case of negativity. Therefore, this study demonstrated that the QuickVue® RSV Test Kit can be effective in early detection of RSV in frozen nasopharyngeal aspirate specimens and is suitable for use as a diagnostic tool in pediatrics.
FundingUniversidade de São Paulo (USP).
Conflicts of interestThe authors declare no conflicts of interest.
The authors would like to thank QUIDEL Corp. for donating the rapid antigen detection tests kits, essential for the development of this work, Gustavo Rezende Melo for the statistical analysis, and Mr. José Maria Lopes for technical support.
Please cite this article as: Mesquita FS, Oliveira DB, Crema D, Pinez CM, Colmanetti TC, Thomazelli LM, et al. Rapid antigen detection test for respiratory syncytial virus diagnosis as a diagnostic tool. J Pediatr (Rio J). 2017;93:246–52.
Study conducted at Universidade de São Paulo, Instituto de Ciências Biomédicas II, Departamento de Microbiologia, Laboratório de Virologia Clínica e Molecular, São Paulo, SP, Brazil.